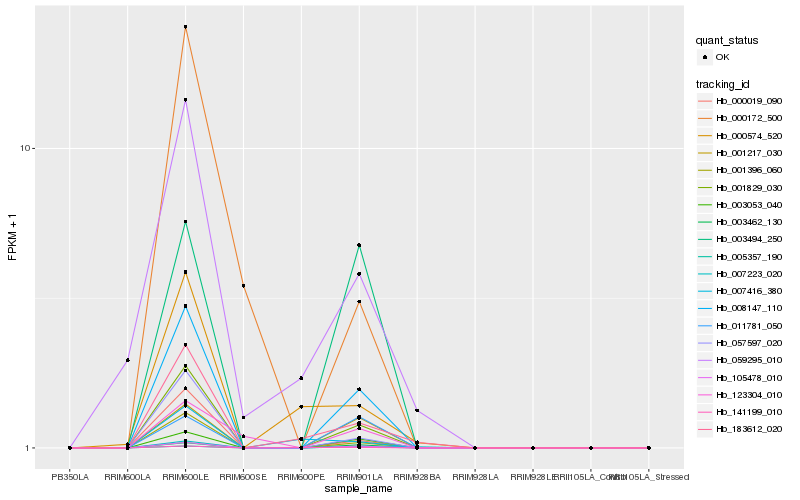
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001829_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 2 |
Hb_008147_110 |
0.0461491505 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_105478_010 |
0.0675761412 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 4 |
Hb_001396_060 |
0.072590263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000019_090 |
0.1099041979 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |
| 6 |
Hb_057597_020 |
0.1147213223 |
- |
- |
PREDICTED: anti-H(O) lectin-like [Gossypium raimondii] |
| 7 |
Hb_003053_040 |
0.1651961241 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003462_130 |
0.1821348695 |
- |
- |
- |
| 9 |
Hb_183612_020 |
0.2012207516 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 10 |
Hb_005357_190 |
0.2023670513 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 11 |
Hb_001217_030 |
0.2032141517 |
- |
- |
- |
| 12 |
Hb_003494_250 |
0.2445735971 |
- |
- |
- |
| 13 |
Hb_000172_500 |
0.2454194184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007223_020 |
0.2459843138 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 15 |
Hb_059295_010 |
0.2526219567 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 16 |
Hb_000574_520 |
0.2603775934 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 17 |
Hb_123304_010 |
0.2810600305 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 18 |
Hb_141199_010 |
0.2812426096 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_007416_380 |
0.2832634406 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 20 |
Hb_011781_050 |
0.2834409586 |
- |
- |
- |