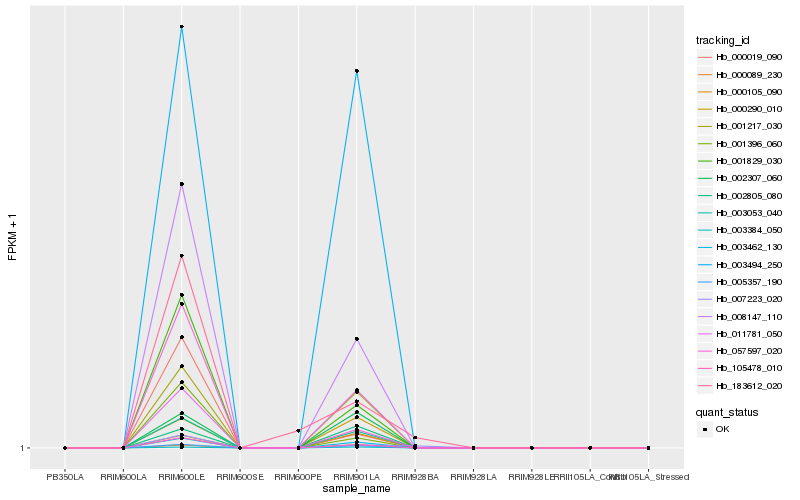
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105478_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 2 |
Hb_008147_110 |
0.0214723833 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_001829_030 |
0.0675761412 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 4 |
Hb_003053_040 |
0.0982383007 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003462_130 |
0.1153400031 |
- |
- |
- |
| 6 |
Hb_005357_190 |
0.1357844487 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 7 |
Hb_001217_030 |
0.1366408339 |
- |
- |
- |
| 8 |
Hb_001396_060 |
0.1395959449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000019_090 |
0.1763116032 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |
| 10 |
Hb_003494_250 |
0.1784895389 |
- |
- |
- |
| 11 |
Hb_057597_020 |
0.181033444 |
- |
- |
PREDICTED: anti-H(O) lectin-like [Gossypium raimondii] |
| 12 |
Hb_011781_050 |
0.2178753369 |
- |
- |
- |
| 13 |
Hb_183612_020 |
0.2224245047 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 14 |
Hb_002307_060 |
0.2309452294 |
- |
- |
- |
| 15 |
Hb_000290_010 |
0.2338582036 |
- |
- |
- |
| 16 |
Hb_000089_230 |
0.2485407327 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 17 |
Hb_003384_050 |
0.2496019998 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_007223_020 |
0.2541915973 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 19 |
Hb_002805_080 |
0.2553685591 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 20 |
Hb_000105_090 |
0.2570864906 |
- |
- |
- |