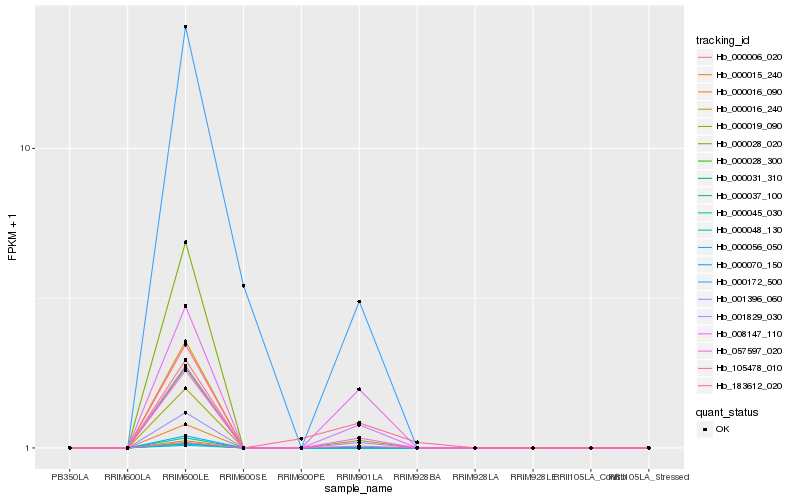
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_090 |
0.0 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |
| 2 |
Hb_057597_020 |
0.004898804 |
- |
- |
PREDICTED: anti-H(O) lectin-like [Gossypium raimondii] |
| 3 |
Hb_001396_060 |
0.0376745926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001829_030 |
0.1099041979 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 5 |
Hb_008147_110 |
0.1553196212 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_105478_010 |
0.1763116032 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 7 |
Hb_000172_500 |
0.2109214965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_183612_020 |
0.2111211192 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 9 |
Hb_000006_020 |
0.2181714593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000015_240 |
0.2181714593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000016_090 |
0.2181714593 |
- |
- |
- |
| 12 |
Hb_000016_240 |
0.2181714593 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0012s13910g [Populus trichocarpa] |
| 13 |
Hb_000028_020 |
0.2181714593 |
- |
- |
- |
| 14 |
Hb_000028_300 |
0.2181714593 |
- |
- |
hypothetical protein 0_13835_01, partial [Pinus radiata] |
| 15 |
Hb_000031_310 |
0.2181714593 |
transcription factor |
TF Family: Orphans |
hypothetical protein [Trichormus azollae] |
| 16 |
Hb_000037_100 |
0.2181714593 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000045_030 |
0.2181714593 |
- |
- |
- |
| 18 |
Hb_000048_130 |
0.2181714593 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000056_050 |
0.2181714593 |
- |
- |
PREDICTED: uncharacterized protein LOC104117826 [Nicotiana tomentosiformis] |
| 20 |
Hb_000070_150 |
0.2181714593 |
- |
- |
hypothetical protein POPTR_0004s19980g [Populus trichocarpa] |