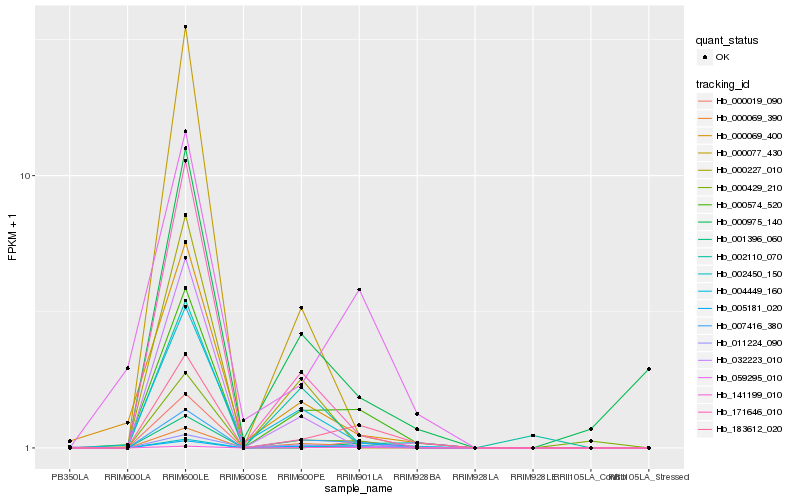
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_520 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007416_380 |
0.1089357732 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 3 |
Hb_000069_390 |
0.1214222537 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 4 |
Hb_183612_020 |
0.1234487519 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 5 |
Hb_002450_150 |
0.1511523748 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_141199_010 |
0.1549703594 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_005181_020 |
0.1871104861 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 8 |
Hb_059295_010 |
0.1877773053 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 9 |
Hb_004449_160 |
0.1932047765 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000069_400 |
0.2071844826 |
- |
- |
RING-H2 finger protein ATL3K, putative [Ricinus communis] |
| 11 |
Hb_000429_210 |
0.214862364 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 12 |
Hb_171646_010 |
0.2188752636 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 13 |
Hb_000975_140 |
0.2373785991 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 14 |
Hb_011224_090 |
0.247944094 |
- |
- |
PREDICTED: ABC transporter G family member 26 [Jatropha curcas] |
| 15 |
Hb_000077_430 |
0.2485434335 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 16 |
Hb_032223_010 |
0.2495554072 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 17 |
Hb_000227_010 |
0.2496282446 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 18 |
Hb_001396_060 |
0.2504616434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002110_070 |
0.2524026857 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000019_090 |
0.2529511701 |
- |
- |
PREDICTED: thioredoxin H4-1-like [Jatropha curcas] |