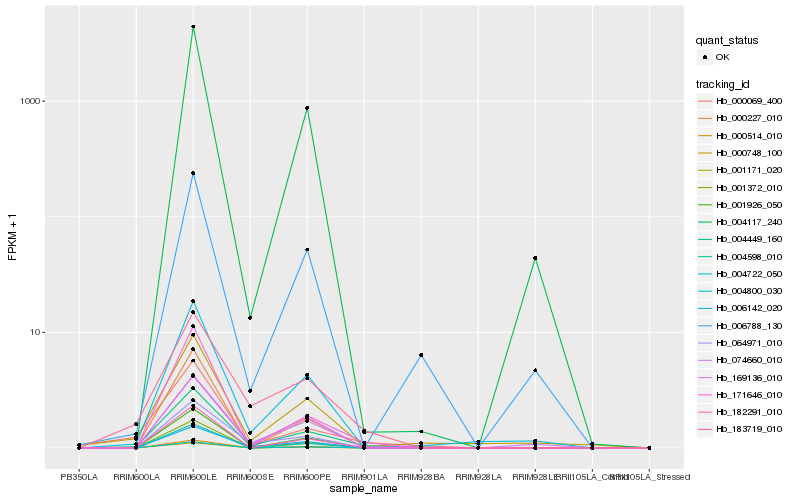
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004449_160 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_004800_030 |
0.1258663208 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 3 |
Hb_074660_010 |
0.1327280841 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 4 |
Hb_171646_010 |
0.1374064709 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 5 |
Hb_169136_010 |
0.1400854906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000069_400 |
0.1416138425 |
- |
- |
RING-H2 finger protein ATL3K, putative [Ricinus communis] |
| 7 |
Hb_000748_100 |
0.1435783895 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 8 |
Hb_006142_020 |
0.146289837 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 9 |
Hb_006788_130 |
0.1474836198 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 10 |
Hb_064971_010 |
0.1482190289 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_000227_010 |
0.1487276059 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 12 |
Hb_004722_050 |
0.1506859577 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 13 |
Hb_000514_010 |
0.1508040846 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 14 |
Hb_004117_240 |
0.1508691089 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 15 |
Hb_001171_020 |
0.1511470424 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 16 |
Hb_183719_010 |
0.1511836415 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 17 |
Hb_001926_050 |
0.1512443282 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 18 |
Hb_001372_010 |
0.1514216365 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_004598_010 |
0.1514548378 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |
| 20 |
Hb_182291_010 |
0.1515848172 |
- |
- |
hypothetical protein CICLE_v10029654mg [Citrus clementina] |