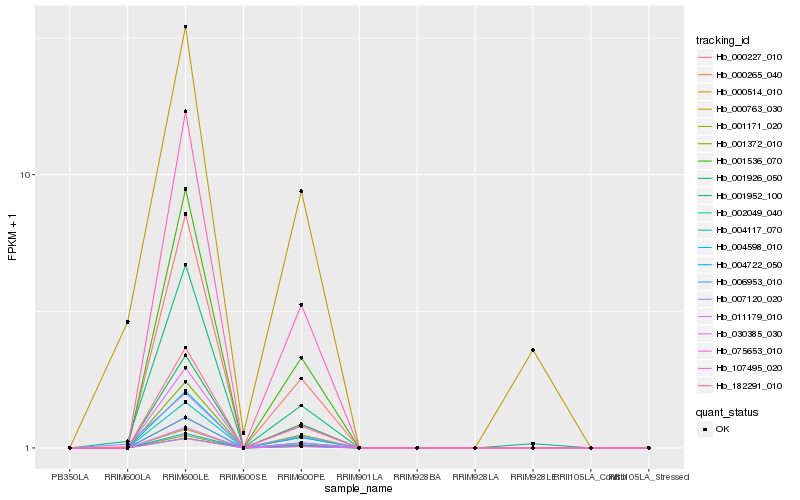
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007120_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001952_100 |
0.1178263325 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 3 |
Hb_000763_030 |
0.1318397565 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000227_010 |
0.1442428659 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 5 |
Hb_004722_050 |
0.1492948114 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 6 |
Hb_001926_050 |
0.1494171525 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 7 |
Hb_000514_010 |
0.1498937823 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 8 |
Hb_004117_070 |
0.149986417 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 9 |
Hb_001171_020 |
0.1504781973 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 10 |
Hb_001372_010 |
0.1508877838 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004598_010 |
0.1509355489 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |
| 12 |
Hb_182291_010 |
0.151119836 |
- |
- |
hypothetical protein CICLE_v10029654mg [Citrus clementina] |
| 13 |
Hb_002049_040 |
0.1512890829 |
- |
- |
PREDICTED: protein UNUSUAL FLORAL ORGANS [Jatropha curcas] |
| 14 |
Hb_006953_010 |
0.1516384011 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_030385_030 |
0.1516552365 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 16 |
Hb_107495_020 |
0.1521516349 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 17 |
Hb_011179_010 |
0.1522894814 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 18 |
Hb_001536_070 |
0.1522957497 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 19 |
Hb_000265_040 |
0.1523184138 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 20 |
Hb_075653_010 |
0.153157792 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |