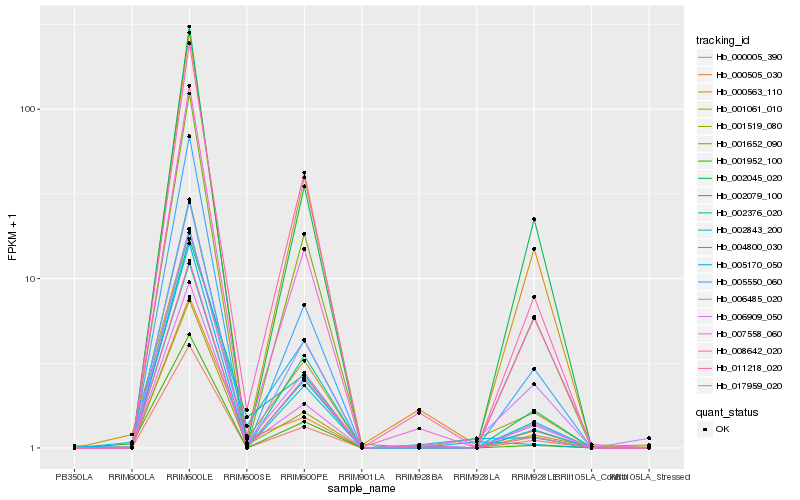
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001061_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100243595 [Vitis vinifera] |
| 2 |
Hb_001519_080 |
0.0988766474 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 3 |
Hb_008642_020 |
0.1030460664 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 4 |
Hb_001952_100 |
0.1039162024 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 5 |
Hb_007558_060 |
0.1057428092 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 6 |
Hb_000005_390 |
0.1058144952 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 7 |
Hb_011218_020 |
0.1076395942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001652_090 |
0.1076696202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000563_110 |
0.108939511 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 10 |
Hb_004800_030 |
0.1094895854 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 11 |
Hb_005550_060 |
0.1129747354 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_005170_050 |
0.1150034047 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 13 |
Hb_000505_030 |
0.1156813259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006485_020 |
0.1156923642 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 15 |
Hb_002843_200 |
0.1157065749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_017959_020 |
0.1197210832 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 17 |
Hb_002079_100 |
0.120232637 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 18 |
Hb_006909_050 |
0.120477693 |
transcription factor |
TF Family: OFP |
hypothetical protein POPTR_0008s01840g [Populus trichocarpa] |
| 19 |
Hb_002045_020 |
0.1211946681 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 20 |
Hb_002376_020 |
0.1212620013 |
- |
- |
lectin, partial [Lathyrus palustris] |