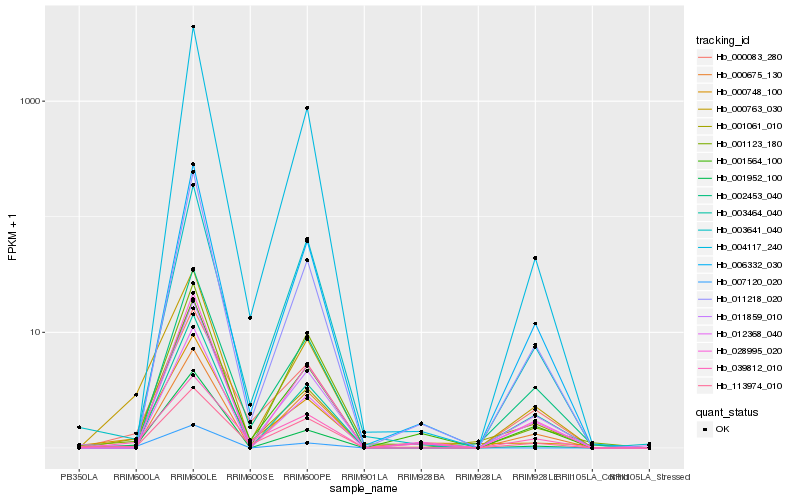
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000763_030 |
0.0 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_007120_020 |
0.1318397565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001061_010 |
0.1406153461 |
- |
- |
PREDICTED: uncharacterized protein LOC100243595 [Vitis vinifera] |
| 4 |
Hb_001952_100 |
0.143862275 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 5 |
Hb_000083_280 |
0.1445738399 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_006332_030 |
0.1498815195 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 7 |
Hb_113974_010 |
0.1525531585 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_011859_010 |
0.1533756907 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 9 |
Hb_039812_010 |
0.1550883649 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_001564_100 |
0.156222693 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 11 |
Hb_011218_020 |
0.1577450075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000675_130 |
0.1595345552 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000748_100 |
0.1600471486 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 14 |
Hb_028995_020 |
0.1601384391 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 15 |
Hb_003641_040 |
0.1603246568 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 16 |
Hb_003464_040 |
0.1626756203 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_012368_040 |
0.1629186971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001123_180 |
0.1631570902 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_004117_240 |
0.1639255515 |
- |
- |
protease inhibitor/seed storage/ltp family [Cicer arietinum] |
| 20 |
Hb_002453_040 |
0.164011964 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |