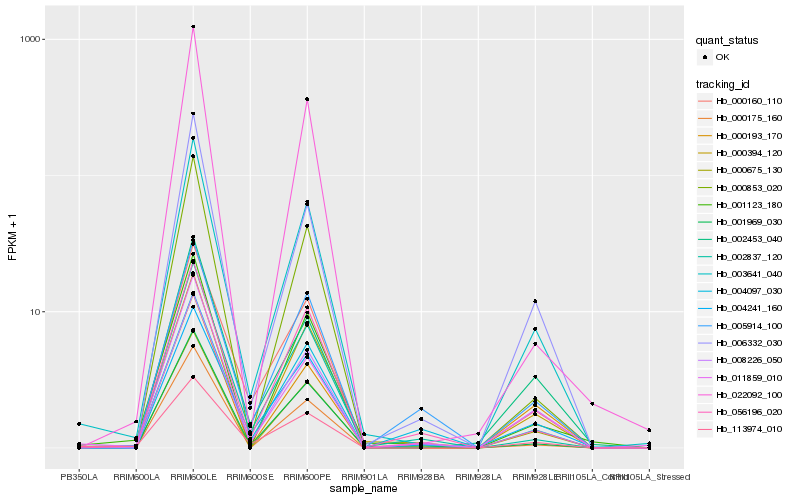
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003641_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 2 |
Hb_000675_130 |
0.0717812848 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001969_030 |
0.0739201081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008226_050 |
0.0763446109 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 5 |
Hb_000853_020 |
0.0820160783 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 6 |
Hb_000394_120 |
0.0825152991 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Populus euphratica] |
| 7 |
Hb_001123_180 |
0.0848135508 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000160_110 |
0.0902157395 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |
| 9 |
Hb_113974_010 |
0.0912689105 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_004241_160 |
0.0927950493 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 11 |
Hb_006332_030 |
0.0942661317 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 12 |
Hb_056196_020 |
0.0956694047 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000175_160 |
0.0958010343 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 14 |
Hb_002837_120 |
0.0974927148 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 15 |
Hb_011859_010 |
0.1038824852 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 16 |
Hb_005914_100 |
0.1062204046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004097_030 |
0.1080702115 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 18 |
Hb_002453_040 |
0.1088757861 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 19 |
Hb_000193_170 |
0.1094792894 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 20 |
Hb_022092_100 |
0.1102329099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |