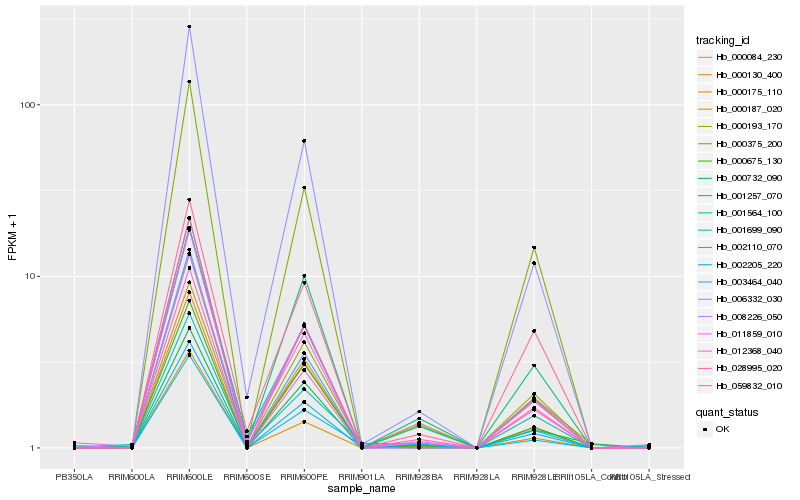
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_220 |
0.0 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 2 |
Hb_000375_200 |
0.0720737381 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 3 |
Hb_028995_020 |
0.0756622733 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 4 |
Hb_000675_130 |
0.0784169929 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_008226_050 |
0.0788652885 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 6 |
Hb_002110_070 |
0.083127231 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000175_110 |
0.084984762 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 8 |
Hb_000193_170 |
0.0853844391 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 9 |
Hb_006332_030 |
0.0862927667 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 10 |
Hb_000130_400 |
0.0870710243 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 11 |
Hb_000732_090 |
0.088526829 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 12 |
Hb_003464_040 |
0.0927829017 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_011859_010 |
0.0929591589 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 14 |
Hb_000187_020 |
0.0949962904 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_001564_100 |
0.0960813006 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 16 |
Hb_012368_040 |
0.0979278762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001257_070 |
0.1002971908 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 18 |
Hb_001699_090 |
0.1021010253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_059832_010 |
0.1023661707 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 20 |
Hb_000084_230 |
0.1062061668 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |