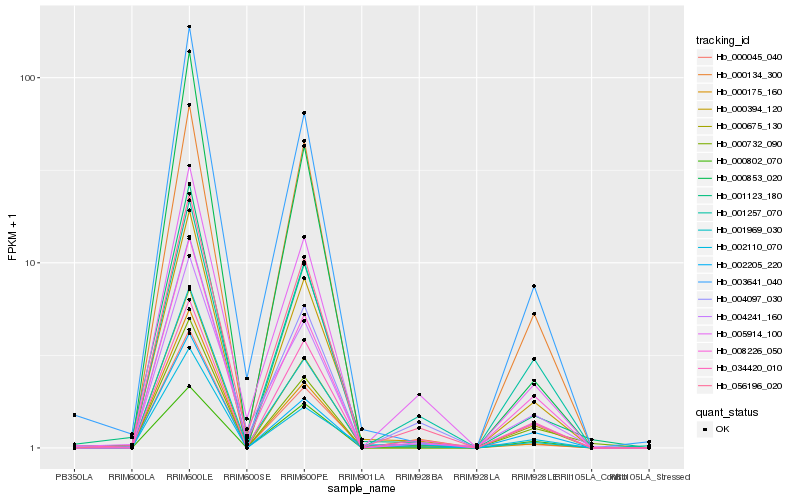
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000394_120 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Populus euphratica] |
| 2 |
Hb_000675_130 |
0.0477273262 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001123_180 |
0.0764975997 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_003641_040 |
0.0825152991 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 5 |
Hb_056196_020 |
0.0918688812 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 6 |
Hb_034420_010 |
0.0931626909 |
- |
- |
glutathione S-transferase [Caragana korshinskii] |
| 7 |
Hb_008226_050 |
0.0958632466 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 8 |
Hb_002110_070 |
0.096099702 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001969_030 |
0.0965669495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000045_040 |
0.0980282831 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 11 |
Hb_000732_090 |
0.101358444 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 12 |
Hb_004097_030 |
0.1057112936 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 13 |
Hb_002205_220 |
0.1063362197 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 14 |
Hb_000853_020 |
0.1077774549 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 15 |
Hb_001257_070 |
0.1081590128 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 16 |
Hb_005914_100 |
0.1082309709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000134_300 |
0.1103939862 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_004241_160 |
0.1155666018 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 19 |
Hb_000175_160 |
0.1157587088 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 20 |
Hb_000802_070 |
0.1186684937 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: eugenol synthase 1-like [Jatropha curcas] |