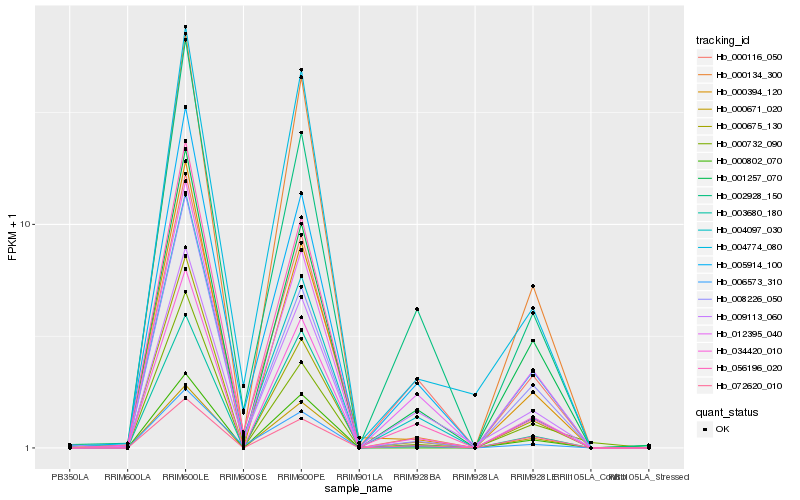
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034420_010 |
0.0 |
- |
- |
glutathione S-transferase [Caragana korshinskii] |
| 2 |
Hb_001257_070 |
0.0685611171 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 3 |
Hb_000134_300 |
0.0777394664 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_012395_040 |
0.082447311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000802_070 |
0.0861653147 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: eugenol synthase 1-like [Jatropha curcas] |
| 6 |
Hb_000671_020 |
0.0917531698 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 7 |
Hb_000394_120 |
0.0931626909 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Populus euphratica] |
| 8 |
Hb_000116_050 |
0.0991550053 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_004774_080 |
0.099172912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003680_180 |
0.1009675124 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 11 |
Hb_006573_310 |
0.1016996655 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 12 |
Hb_009113_060 |
0.1048129409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005914_100 |
0.1053167089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_072620_010 |
0.107559926 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 15 |
Hb_000732_090 |
0.1078935974 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 16 |
Hb_002928_150 |
0.1092580135 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 17 |
Hb_004097_030 |
0.1103654832 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 18 |
Hb_000675_130 |
0.1108323516 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_008226_050 |
0.1112074222 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 20 |
Hb_056196_020 |
0.1140924073 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |