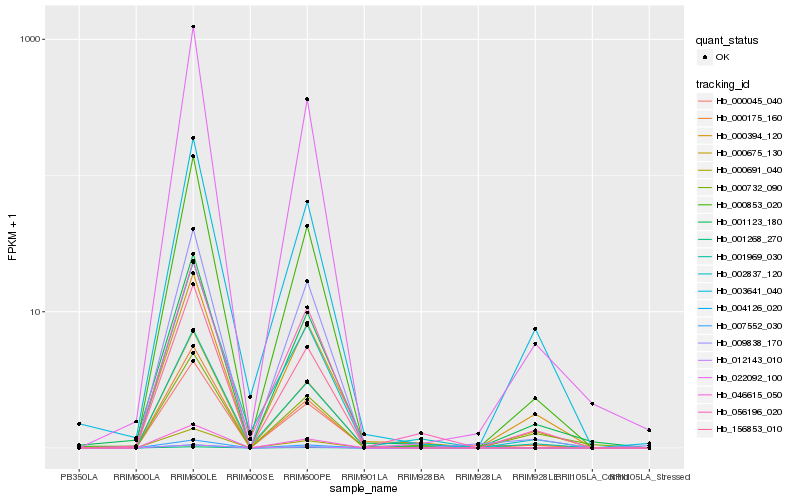
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_180 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000394_120 |
0.0764975997 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Populus euphratica] |
| 3 |
Hb_000853_020 |
0.0824354707 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 4 |
Hb_000675_130 |
0.0832821811 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001969_030 |
0.0837805088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003641_040 |
0.0848135508 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 7 |
Hb_000175_160 |
0.0926688886 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 8 |
Hb_022092_100 |
0.0929589001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000045_040 |
0.0957793767 |
- |
- |
anthranilate synthase component I, putative [Ricinus communis] |
| 10 |
Hb_056196_020 |
0.0979629694 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009838_170 |
0.104711367 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 12 |
Hb_002837_120 |
0.1096727634 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 13 |
Hb_156853_010 |
0.1143949165 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 14 |
Hb_004126_020 |
0.1159738019 |
- |
- |
PREDICTED: uncharacterized protein LOC105649549 [Jatropha curcas] |
| 15 |
Hb_001268_270 |
0.1159784632 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000691_040 |
0.1159867934 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 17 |
Hb_046615_050 |
0.1160100389 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 18 |
Hb_000732_090 |
0.1160524178 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 19 |
Hb_007552_030 |
0.1161395734 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 20 |
Hb_012143_010 |
0.1161530702 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Vitis vinifera] |