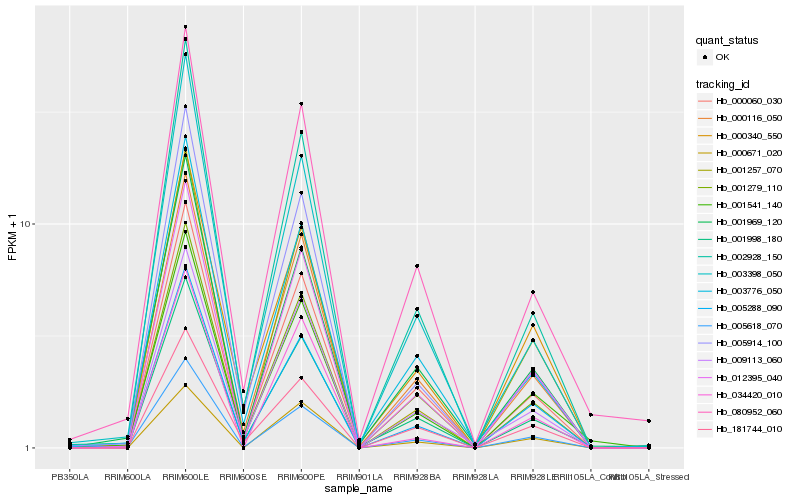
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000116_050 |
0.048208122 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001279_110 |
0.0508290721 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 4 |
Hb_005618_070 |
0.0531114073 |
- |
- |
PREDICTED: uncharacterized protein LOC105643318 [Jatropha curcas] |
| 5 |
Hb_001257_070 |
0.0615002483 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 6 |
Hb_000060_030 |
0.0615484486 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |
| 7 |
Hb_005288_090 |
0.0617040631 |
- |
- |
PREDICTED: uncharacterized protein LOC105647753 [Jatropha curcas] |
| 8 |
Hb_001541_140 |
0.0627927626 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 9 |
Hb_002928_150 |
0.0632306793 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 10 |
Hb_009113_060 |
0.067244816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001969_120 |
0.0726169164 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 12 |
Hb_003776_050 |
0.0782062581 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 13 |
Hb_000340_550 |
0.0794474654 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 14 |
Hb_001998_180 |
0.0796104352 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 15 |
Hb_005914_100 |
0.0822417248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_034420_010 |
0.082447311 |
- |
- |
glutathione S-transferase [Caragana korshinskii] |
| 17 |
Hb_181744_010 |
0.084234477 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_080952_060 |
0.0858651203 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 19 |
Hb_003398_050 |
0.0876930144 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 20 |
Hb_000671_020 |
0.0884913872 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |