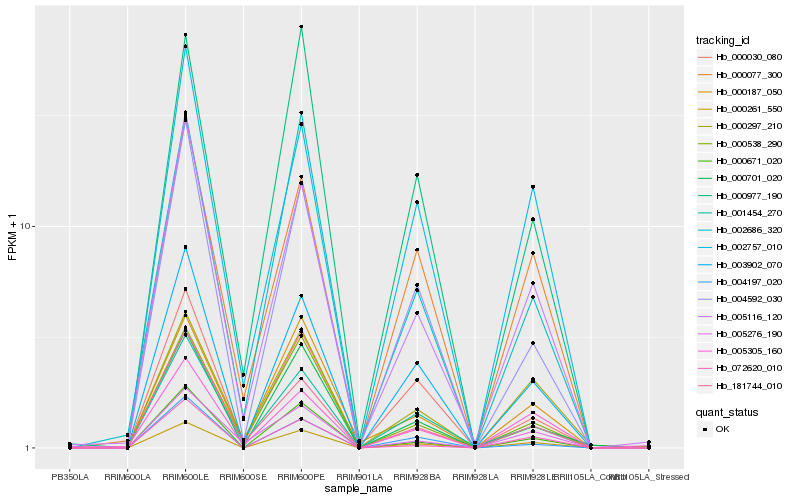
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_550 |
0.0 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 2 |
Hb_003902_070 |
0.0759134727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000671_020 |
0.0873665351 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 4 |
Hb_005116_120 |
0.1015493789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000701_020 |
0.1071456719 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004592_030 |
0.1126557034 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_002757_010 |
0.1129343378 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 8 |
Hb_000077_300 |
0.113118951 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 9 |
Hb_072620_010 |
0.1142028586 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Cucumis melo] |
| 10 |
Hb_001454_270 |
0.1203282898 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000297_210 |
0.1220079428 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 12 |
Hb_002686_320 |
0.124794941 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_000187_050 |
0.1257055887 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 14 |
Hb_000538_290 |
0.1267390048 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 15 |
Hb_004197_020 |
0.1277495772 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 16 |
Hb_181744_010 |
0.1279270365 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 17 |
Hb_000030_080 |
0.1298938679 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 18 |
Hb_000977_190 |
0.1320213339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005305_160 |
0.1340353704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005276_190 |
0.1342346766 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |