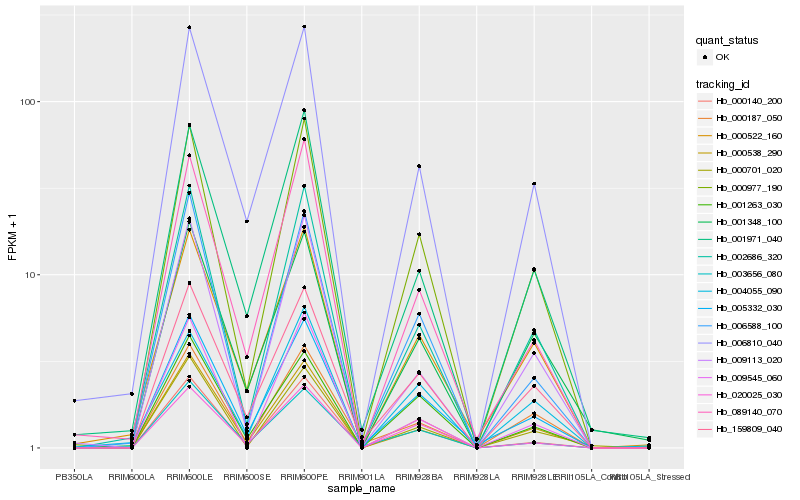
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002686_320 |
0.0660353293 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 3 |
Hb_005332_030 |
0.0801781727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_159809_040 |
0.0902582883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000538_290 |
0.0915372245 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_000140_200 |
0.0920539937 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 7 |
Hb_020025_030 |
0.0921508474 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 8 |
Hb_000701_020 |
0.0945894432 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000187_050 |
0.0947479347 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 10 |
Hb_009113_020 |
0.0949987579 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 11 |
Hb_006810_040 |
0.095200927 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_006588_100 |
0.0954832948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000522_160 |
0.1017038554 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 14 |
Hb_001263_030 |
0.1048390541 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 15 |
Hb_089140_070 |
0.1057704325 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 16 |
Hb_001971_040 |
0.1060778448 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 17 |
Hb_003656_080 |
0.1083426378 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 18 |
Hb_004055_090 |
0.10863551 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 19 |
Hb_009545_060 |
0.1089653258 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 20 |
Hb_001348_100 |
0.1180465414 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |