| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
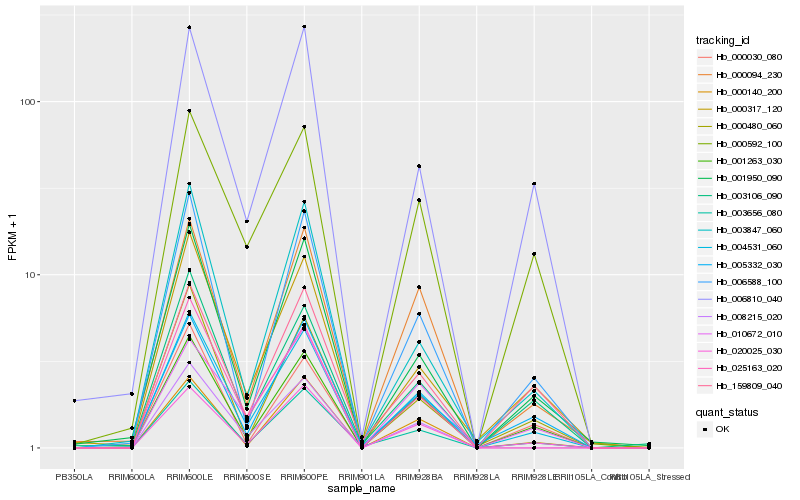
Hb_004531_060 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_001263_030 |
0.0769600303 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 3 |
Hb_003656_080 |
0.0791660401 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 4 |
Hb_020025_030 |
0.0839475163 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 5 |
Hb_000140_200 |
0.099491354 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 6 |
Hb_006588_100 |
0.1003936204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001950_090 |
0.101562912 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 8 |
Hb_005332_030 |
0.103583549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000094_230 |
0.1043250374 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 10 |
Hb_008215_020 |
0.1063111862 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000317_120 |
0.1063582383 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 12 |
Hb_003106_090 |
0.1119573731 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 13 |
Hb_006810_040 |
0.1128453886 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_003847_060 |
0.1134103293 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_025163_020 |
0.1146210719 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 16 |
Hb_000592_100 |
0.1168956113 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 17 |
Hb_000480_060 |
0.1183991856 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 18 |
Hb_159809_040 |
0.1185997092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000030_080 |
0.1218078622 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 20 |
Hb_010672_010 |
0.1218630328 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |