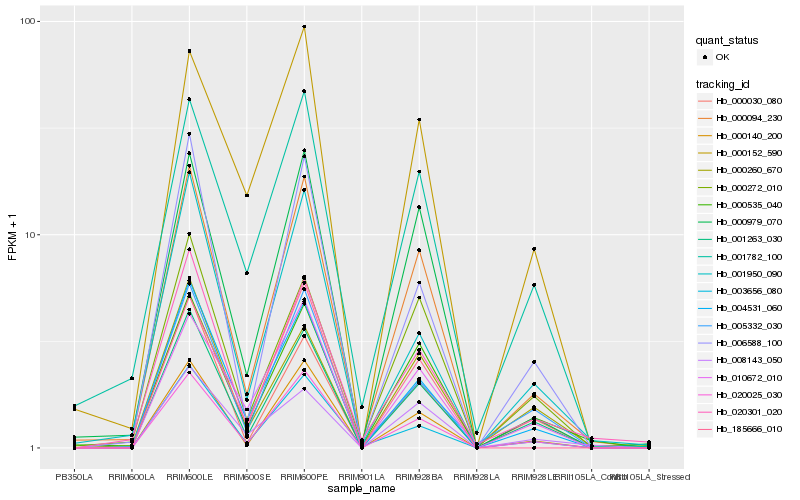
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_230 |
0.0 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 2 |
Hb_000979_070 |
0.0792539908 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 3 |
Hb_000535_040 |
0.08001822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 4 |
Hb_020025_030 |
0.0821583223 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 5 |
Hb_000140_200 |
0.0847832692 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 6 |
Hb_001263_030 |
0.0967198523 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 7 |
Hb_004531_060 |
0.1043250374 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 8 |
Hb_020301_020 |
0.1090780255 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_003656_080 |
0.1120312111 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 10 |
Hb_000272_010 |
0.1180393096 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 11 |
Hb_006588_100 |
0.1217136573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005332_030 |
0.1241163839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001782_100 |
0.1253495052 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 14 |
Hb_185666_010 |
0.1270468377 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 15 |
Hb_010672_010 |
0.1301546107 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 16 |
Hb_001950_090 |
0.134272025 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 17 |
Hb_000030_080 |
0.1346855207 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 18 |
Hb_008143_050 |
0.137733296 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 19 |
Hb_000260_670 |
0.1416777632 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 20 |
Hb_000152_590 |
0.1435747581 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |