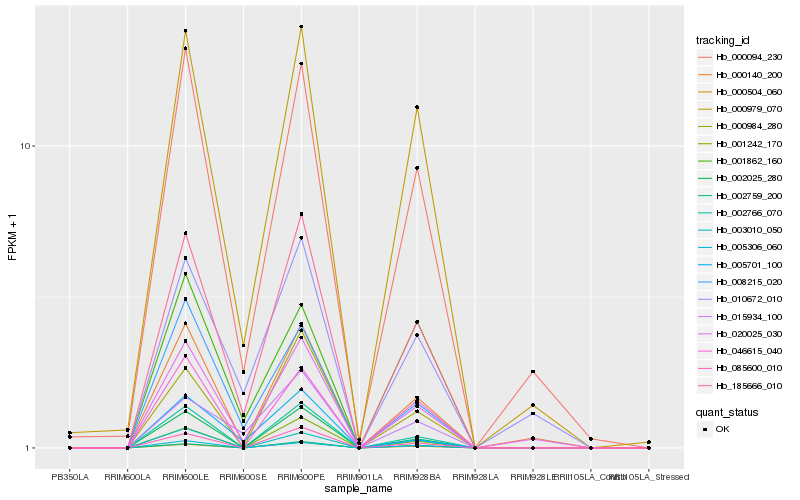
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185666_010 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_000979_070 |
0.1062124774 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 3 |
Hb_015934_100 |
0.1126904745 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 4 |
Hb_020025_030 |
0.1173559362 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 5 |
Hb_008215_020 |
0.1181396436 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000504_060 |
0.1248656525 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 7 |
Hb_000094_230 |
0.1270468377 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 8 |
Hb_000140_200 |
0.1279832874 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 9 |
Hb_085600_010 |
0.129899655 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 10 |
Hb_002766_070 |
0.1309032097 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 11 |
Hb_002759_200 |
0.1333941496 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 12 |
Hb_002025_280 |
0.1371294249 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 13 |
Hb_001242_170 |
0.137159844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005701_100 |
0.137222393 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 15 |
Hb_000984_280 |
0.1389779427 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 16 |
Hb_046615_040 |
0.141178752 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_010672_010 |
0.1434462643 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 18 |
Hb_005306_060 |
0.1450887149 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 19 |
Hb_003010_050 |
0.1455001109 |
- |
- |
- |
| 20 |
Hb_001862_160 |
0.1457053964 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |