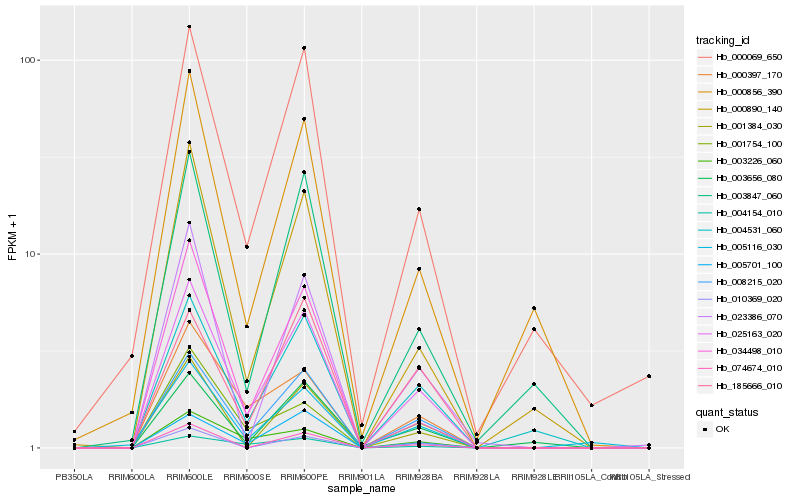
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025163_020 |
0.0 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 2 |
Hb_008215_020 |
0.0320378866 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_004531_060 |
0.1146210719 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 4 |
Hb_005701_100 |
0.1232682199 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_000890_140 |
0.1242678698 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 6 |
Hb_005116_030 |
0.1280795116 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000397_170 |
0.128705214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000069_650 |
0.1295516096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_034498_010 |
0.1334121423 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 10 |
Hb_003847_060 |
0.1403464108 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_003226_060 |
0.1420566979 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 12 |
Hb_003656_080 |
0.1443796408 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 13 |
Hb_074674_010 |
0.1445468621 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 14 |
Hb_001754_100 |
0.1451789078 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 15 |
Hb_001384_030 |
0.147685197 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 16 |
Hb_023386_070 |
0.1494044877 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 17 |
Hb_185666_010 |
0.1496331821 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 18 |
Hb_010369_020 |
0.1513212448 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 19 |
Hb_000856_390 |
0.1514520026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004154_010 |
0.1515295465 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |