| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
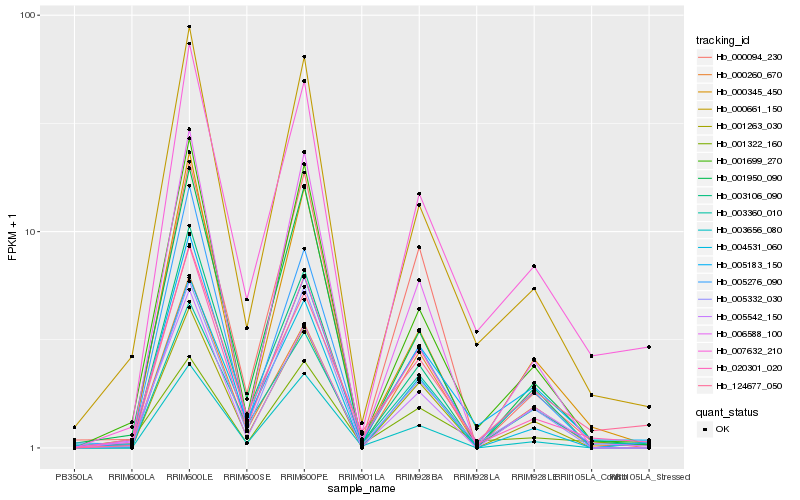
Hb_020301_020 |
0.0 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 2 |
Hb_000661_150 |
0.0981358844 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 3 |
Hb_005183_150 |
0.1033491907 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 4 |
Hb_001699_270 |
0.1076731029 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 5 |
Hb_007632_210 |
0.1080360247 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 6 |
Hb_000094_230 |
0.1090780255 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 7 |
Hb_005542_150 |
0.1091754812 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 8 |
Hb_003360_010 |
0.1112901374 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 9 |
Hb_003656_080 |
0.1168985068 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 10 |
Hb_001950_090 |
0.1179743744 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 11 |
Hb_124677_050 |
0.1196495719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000260_670 |
0.1198800718 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 13 |
Hb_005332_030 |
0.1210550842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001322_160 |
0.1232206058 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 15 |
Hb_003106_090 |
0.1238038429 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 16 |
Hb_006588_100 |
0.1242589628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001263_030 |
0.1242964884 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 18 |
Hb_000345_450 |
0.1246524006 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 19 |
Hb_004531_060 |
0.1251539327 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 20 |
Hb_005276_090 |
0.1262452185 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |