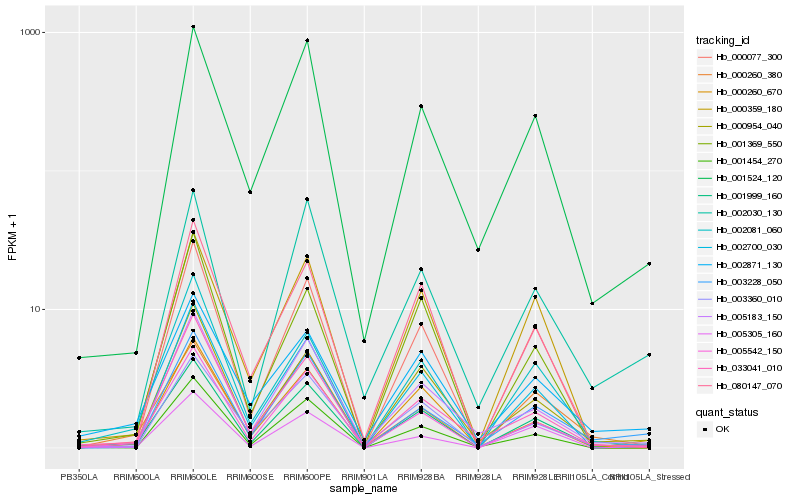
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 2 |
Hb_005542_150 |
0.0896698732 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 3 |
Hb_002081_060 |
0.1106421826 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 4 |
Hb_000954_040 |
0.1126560508 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 5 |
Hb_000260_670 |
0.1135883499 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 6 |
Hb_002700_030 |
0.113654237 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 7 |
Hb_003228_050 |
0.1146630855 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 8 |
Hb_000077_300 |
0.1163867315 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 9 |
Hb_005305_160 |
0.1170397574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000359_180 |
0.1208185818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001524_120 |
0.124805856 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_002871_130 |
0.1291619628 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 13 |
Hb_080147_070 |
0.1293250824 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 14 |
Hb_033041_010 |
0.1301116434 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 15 |
Hb_003360_010 |
0.1305384663 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 16 |
Hb_005183_150 |
0.1326936961 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 17 |
Hb_000260_380 |
0.1332208875 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 18 |
Hb_001369_550 |
0.1335138455 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 19 |
Hb_002030_130 |
0.1367935158 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 20 |
Hb_001454_270 |
0.1368006827 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |