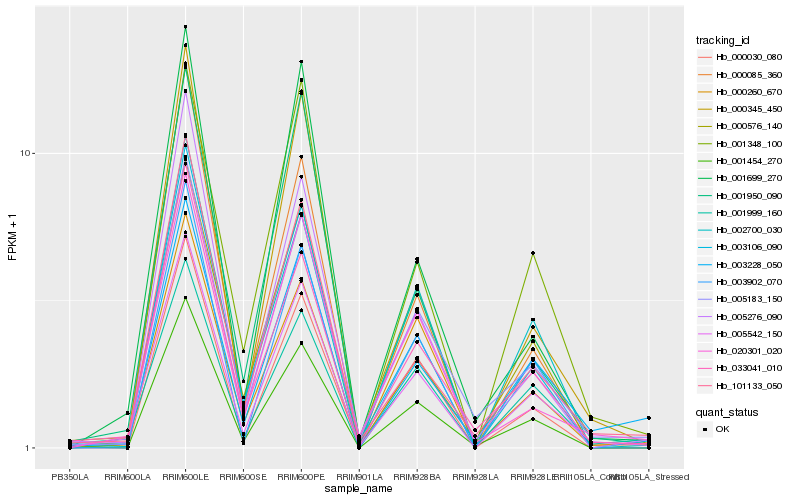
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 2 |
Hb_003106_090 |
0.0862856059 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 3 |
Hb_001999_160 |
0.0896698732 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 4 |
Hb_000345_450 |
0.1001094098 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 5 |
Hb_033041_010 |
0.1016329639 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 6 |
Hb_000085_360 |
0.1017908831 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 7 |
Hb_101133_050 |
0.1038196087 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 8 |
Hb_001454_270 |
0.1065152236 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005183_150 |
0.1069830239 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 10 |
Hb_001348_100 |
0.1082690382 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 11 |
Hb_020301_020 |
0.1091754812 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_000576_140 |
0.1101576857 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 13 |
Hb_002700_030 |
0.1119094647 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 14 |
Hb_003228_050 |
0.1132417019 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 15 |
Hb_000030_080 |
0.1141928651 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 16 |
Hb_000260_670 |
0.1149222429 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 17 |
Hb_003902_070 |
0.1159799325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001950_090 |
0.1162164051 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 19 |
Hb_005276_090 |
0.1184115216 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001699_270 |
0.1187976083 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |