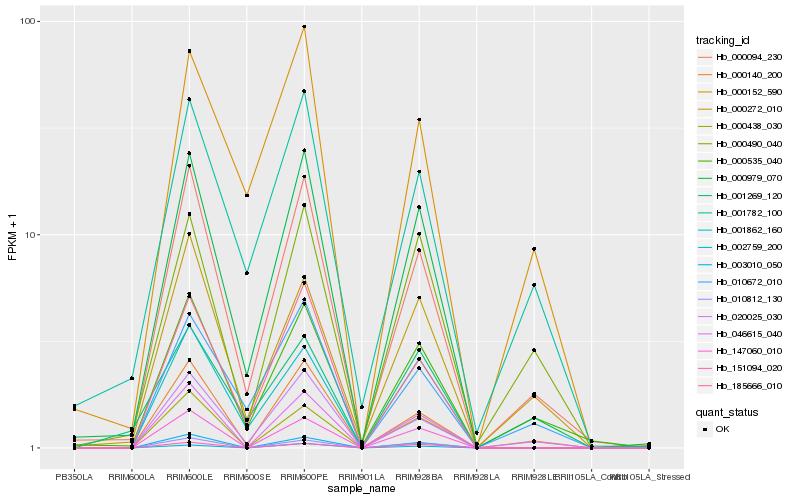
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_070 |
0.0 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 2 |
Hb_000094_230 |
0.0792539908 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 3 |
Hb_185666_010 |
0.1062124774 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 4 |
Hb_000535_040 |
0.1170084415 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 5 |
Hb_001862_160 |
0.1183535015 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 6 |
Hb_020025_030 |
0.1227643192 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 7 |
Hb_000140_200 |
0.1265629405 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 8 |
Hb_046615_040 |
0.1342860164 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_010812_130 |
0.1370380639 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 10 |
Hb_147060_010 |
0.1380005542 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 11 |
Hb_003010_050 |
0.1428413482 |
- |
- |
- |
| 12 |
Hb_010672_010 |
0.1431766658 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 13 |
Hb_000272_010 |
0.143444168 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 14 |
Hb_151094_020 |
0.1442909445 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 15 |
Hb_001782_100 |
0.1445431731 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 16 |
Hb_000438_030 |
0.1472365333 |
- |
- |
- |
| 17 |
Hb_002759_200 |
0.1487464233 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 18 |
Hb_000152_590 |
0.149521337 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 19 |
Hb_001269_120 |
0.150150255 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 20 |
Hb_000490_040 |
0.1507485091 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |