| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
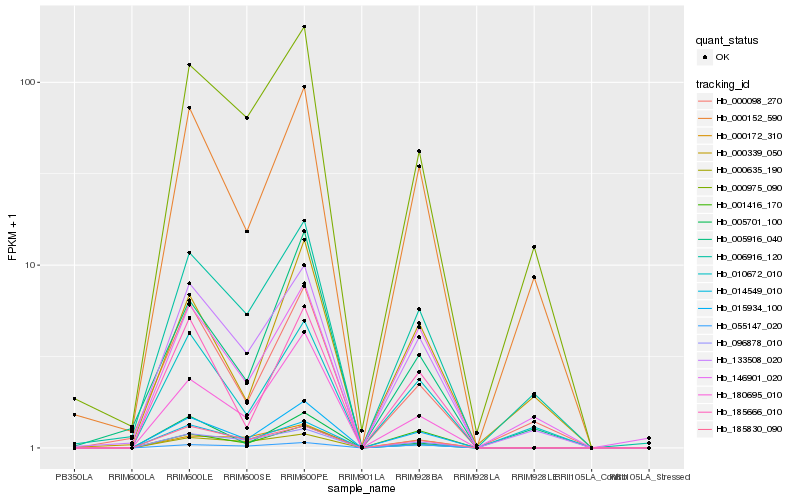
Hb_015934_100 |
0.0 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 2 |
Hb_180695_010 |
0.0910190306 |
- |
- |
hypothetical protein CICLE_v10013449mg [Citrus clementina] |
| 3 |
Hb_185666_010 |
0.1126904745 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 4 |
Hb_133508_020 |
0.1159481879 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 5 |
Hb_096878_010 |
0.125875259 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 6 |
Hb_185830_090 |
0.1309271953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010672_010 |
0.1442391849 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 8 |
Hb_055147_020 |
0.1461553035 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 9 |
Hb_000152_590 |
0.1482794284 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_006916_120 |
0.1490210911 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 11 |
Hb_000635_190 |
0.1498684202 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 12 |
Hb_005916_040 |
0.1503454957 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 13 |
Hb_005701_100 |
0.1582327343 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 14 |
Hb_000172_310 |
0.1591601829 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 15 |
Hb_146901_020 |
0.1615120879 |
- |
- |
- |
| 16 |
Hb_014549_010 |
0.1634016199 |
- |
- |
PREDICTED: uncharacterized protein LOC101512634 [Cicer arietinum] |
| 17 |
Hb_000098_270 |
0.1640057769 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 18 |
Hb_001416_170 |
0.164867914 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000975_090 |
0.1672310207 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 20 |
Hb_000339_050 |
0.1679760511 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |