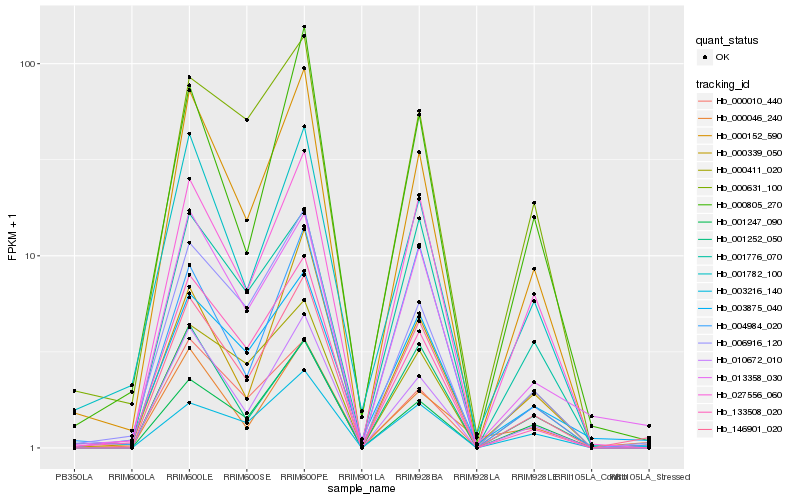
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146901_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000152_590 |
0.0979571325 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 3 |
Hb_027556_060 |
0.1011827266 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 4 |
Hb_010672_010 |
0.1019630299 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 5 |
Hb_013358_030 |
0.1059161691 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 6 |
Hb_003875_040 |
0.1152636101 |
- |
- |
PREDICTED: endoglucanase 11-like [Jatropha curcas] |
| 7 |
Hb_003216_140 |
0.1214418828 |
- |
- |
F-box protein SNE, putative [Ricinus communis] |
| 8 |
Hb_133508_020 |
0.1242361294 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_006916_120 |
0.1248367165 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 10 |
Hb_001776_070 |
0.1319440843 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 11 |
Hb_001247_090 |
0.13345983 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 12 |
Hb_004984_020 |
0.1337135749 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 13 |
Hb_000411_020 |
0.142882076 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001782_100 |
0.1442802143 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_000805_270 |
0.1461879096 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 16 |
Hb_000339_050 |
0.1485194805 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 17 |
Hb_000631_100 |
0.1486714618 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 18 |
Hb_001252_050 |
0.1493267208 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 19 |
Hb_000046_240 |
0.1513720427 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 20 |
Hb_000010_440 |
0.1518204703 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |