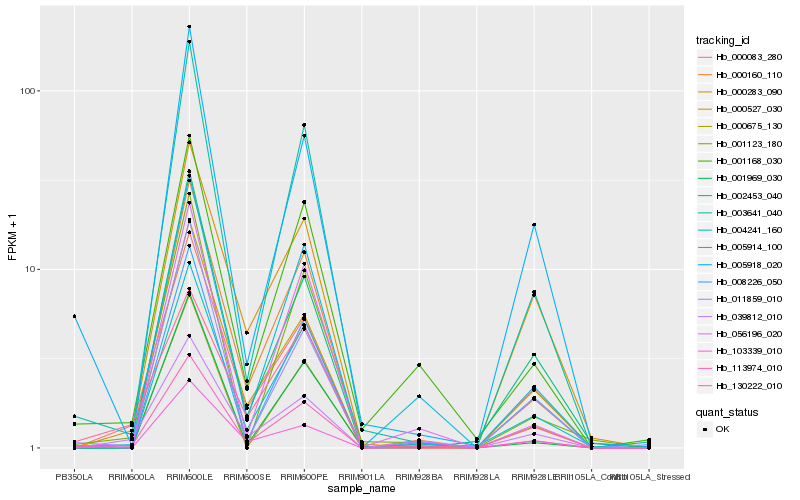
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113974_010 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_003641_040 |
0.0912689105 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 3 |
Hb_000160_110 |
0.0948315961 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |
| 4 |
Hb_004241_160 |
0.1075518118 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 5 |
Hb_000083_280 |
0.1077018869 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_008226_050 |
0.1147205625 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 7 |
Hb_000527_030 |
0.1167746325 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 8 |
Hb_005918_020 |
0.1226380717 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 9 |
Hb_001168_030 |
0.1292479001 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 10 |
Hb_002453_040 |
0.130266593 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 11 |
Hb_005914_100 |
0.1310093137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_130222_010 |
0.1321380388 |
- |
- |
hypothetical protein RCOM_1272620 [Ricinus communis] |
| 13 |
Hb_103339_010 |
0.1331326106 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000675_130 |
0.1335492347 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000283_090 |
0.1350566954 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_011859_010 |
0.1352268663 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 17 |
Hb_001123_180 |
0.1354316139 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001969_030 |
0.1383658612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_056196_020 |
0.1388949882 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 20 |
Hb_039812_010 |
0.1390189459 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |