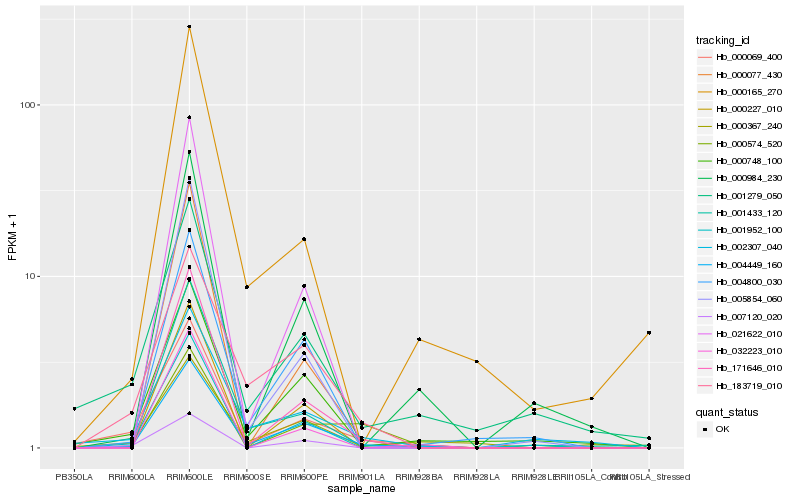
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_400 |
0.0 |
- |
- |
RING-H2 finger protein ATL3K, putative [Ricinus communis] |
| 2 |
Hb_004449_160 |
0.1416138425 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001279_050 |
0.1535668966 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 4 |
Hb_002307_040 |
0.1654313864 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_007120_020 |
0.1728644966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000748_100 |
0.1729868472 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 7 |
Hb_171646_010 |
0.1852465748 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 8 |
Hb_001952_100 |
0.1863436077 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 9 |
Hb_183719_010 |
0.1959179124 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 10 |
Hb_001433_120 |
0.1968477916 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 11 |
Hb_000367_240 |
0.202549043 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 12 |
Hb_021622_010 |
0.2068289273 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 13 |
Hb_000574_520 |
0.2071844826 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 14 |
Hb_032223_010 |
0.2073738839 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 15 |
Hb_000077_430 |
0.2081181787 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 16 |
Hb_000227_010 |
0.2087208118 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 17 |
Hb_005854_060 |
0.210871965 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_004800_030 |
0.2114758608 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 19 |
Hb_000984_230 |
0.211525402 |
- |
- |
- |
| 20 |
Hb_000165_270 |
0.2121939126 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |