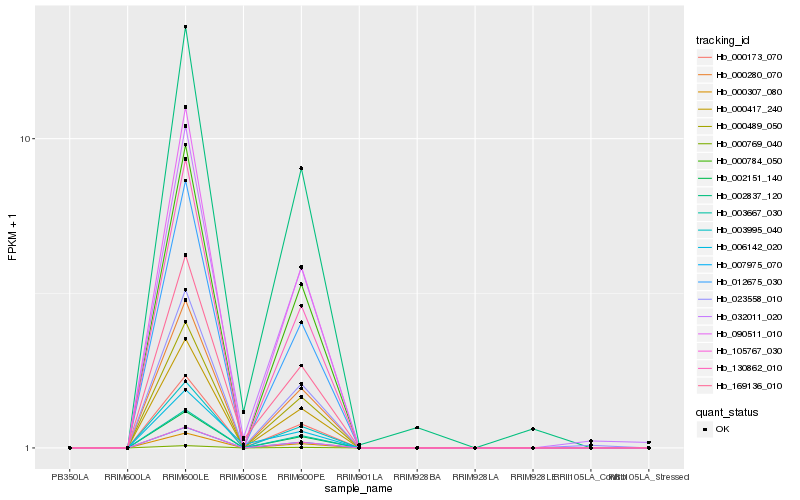
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169136_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006142_020 |
0.0616857023 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 3 |
Hb_032011_020 |
0.0738087846 |
- |
- |
alpha-farnesene synthase [Ricinus communis] |
| 4 |
Hb_002837_120 |
0.084828061 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 5 |
Hb_023558_010 |
0.0856329486 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 6 |
Hb_007975_070 |
0.0890400942 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 7 |
Hb_105767_030 |
0.089051207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000417_240 |
0.0891758 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 9 |
Hb_003995_040 |
0.0891836058 |
- |
- |
PREDICTED: tropinone reductase homolog At2g29290-like [Jatropha curcas] |
| 10 |
Hb_000173_070 |
0.0892246058 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 11 |
Hb_000307_080 |
0.0892935438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000280_070 |
0.0894618767 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_000784_050 |
0.0895140714 |
- |
- |
PREDICTED: auxin-induced protein X10A-like [Nelumbo nucifera] |
| 14 |
Hb_130862_010 |
0.0896723104 |
- |
- |
purine permease, putative [Ricinus communis] |
| 15 |
Hb_002151_140 |
0.0897075763 |
- |
- |
PREDICTED: lichenase-2-like [Pyrus x bretschneideri] |
| 16 |
Hb_090511_010 |
0.0899655994 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_012675_030 |
0.0900098718 |
- |
- |
hypothetical protein POPTR_0013s04780g [Populus trichocarpa] |
| 18 |
Hb_003667_030 |
0.090770503 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL15 [Jatropha curcas] |
| 19 |
Hb_000769_040 |
0.0908301028 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 20 |
Hb_000489_050 |
0.091198114 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |