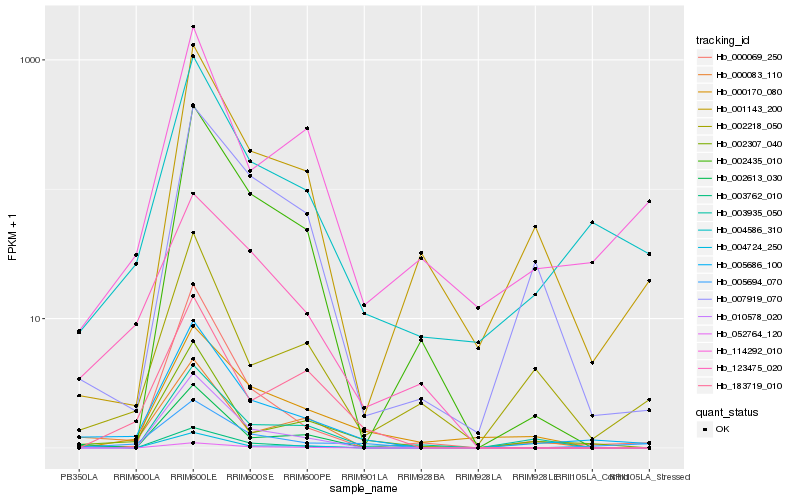
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005686_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004586_310 |
0.1237175806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005694_070 |
0.1556168922 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_002307_040 |
0.1568321884 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000069_250 |
0.1892349589 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 6 |
Hb_114292_010 |
0.1922438579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003762_010 |
0.1939581378 |
- |
- |
- |
| 8 |
Hb_002613_030 |
0.1962071488 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001143_200 |
0.2015704287 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 10 |
Hb_010578_020 |
0.2017021354 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003935_050 |
0.2057295306 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_007919_070 |
0.2065842029 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 13 |
Hb_052764_120 |
0.2079078047 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 14 |
Hb_123475_020 |
0.2088647754 |
- |
- |
- |
| 15 |
Hb_000170_080 |
0.2089063903 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 16 |
Hb_000083_110 |
0.2099191092 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 17 |
Hb_002218_050 |
0.2113140327 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 18 |
Hb_002435_010 |
0.2120064343 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 19 |
Hb_004724_250 |
0.213065548 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_183719_010 |
0.2131461363 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |