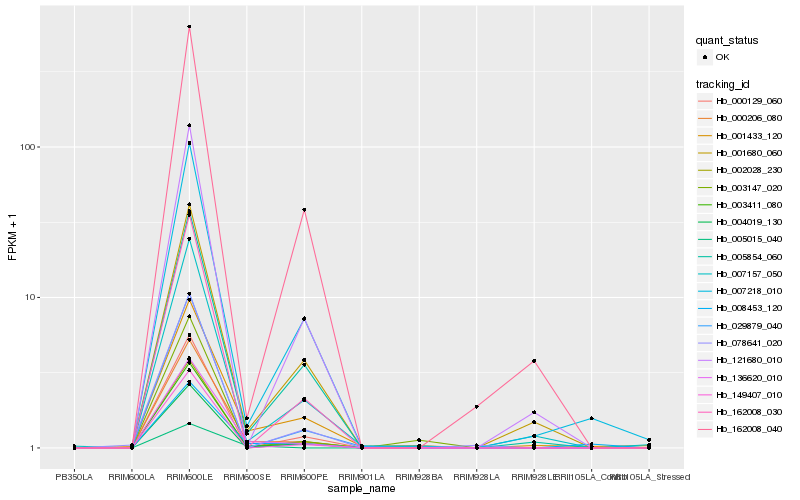
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136620_010 |
0.0 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 2 |
Hb_000206_080 |
0.024558805 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 3 |
Hb_003411_080 |
0.0768357444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002028_230 |
0.1072122145 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 5 |
Hb_005854_060 |
0.1281247721 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001433_120 |
0.1285558028 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 7 |
Hb_149407_010 |
0.128680417 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 8 |
Hb_008453_120 |
0.1297039792 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 9 |
Hb_005015_040 |
0.1299618646 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1-like [Nicotiana sylvestris] |
| 10 |
Hb_162008_030 |
0.1306931256 |
- |
- |
cytochrome p450 [Ageratina adenophora] |
| 11 |
Hb_000129_060 |
0.1346338003 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 12 |
Hb_029879_040 |
0.1347412824 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 13 |
Hb_007157_050 |
0.1352644398 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 14 |
Hb_004019_130 |
0.1363980706 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 15 |
Hb_007218_010 |
0.1390571194 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 16 |
Hb_001680_060 |
0.141665157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003147_020 |
0.1428429119 |
- |
- |
hypothetical protein POPTR_0019s07690g [Populus trichocarpa] |
| 18 |
Hb_121680_010 |
0.1449160904 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 19 |
Hb_078641_020 |
0.1461661306 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_162008_040 |
0.1483706446 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |