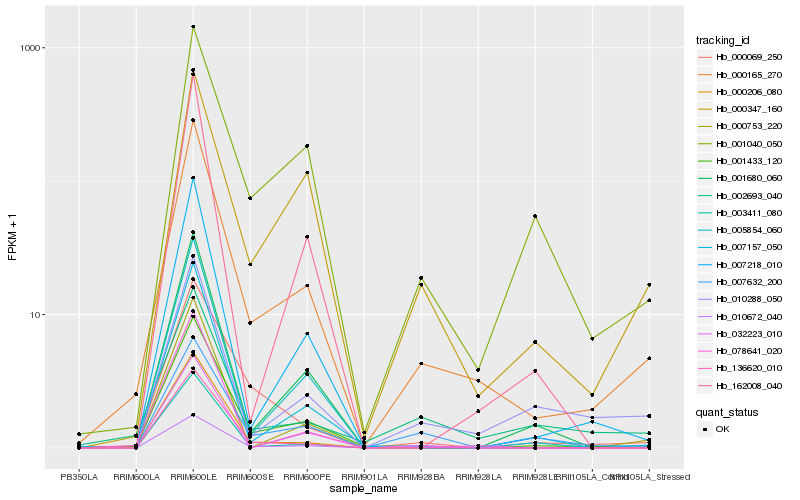
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_270 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 2 |
Hb_001433_120 |
0.093563032 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 3 |
Hb_007218_010 |
0.1430894757 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 4 |
Hb_010672_040 |
0.1462896073 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_007157_050 |
0.146872025 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 6 |
Hb_000347_160 |
0.1496517535 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 7 |
Hb_005854_060 |
0.1533099059 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000206_080 |
0.1549090211 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 9 |
Hb_136620_010 |
0.155191244 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 10 |
Hb_001040_050 |
0.155954063 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_001680_060 |
0.1592287677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010288_050 |
0.1615863016 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 13 |
Hb_000753_220 |
0.1627805347 |
- |
- |
hypothetical protein RCOM_1467930 [Ricinus communis] |
| 14 |
Hb_003411_080 |
0.1632508996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000069_250 |
0.1635045454 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 16 |
Hb_162008_040 |
0.1642237831 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 17 |
Hb_002693_040 |
0.1643863934 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 18 |
Hb_007632_200 |
0.1648632001 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 19 |
Hb_032223_010 |
0.166864616 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 20 |
Hb_078641_020 |
0.1668832567 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |