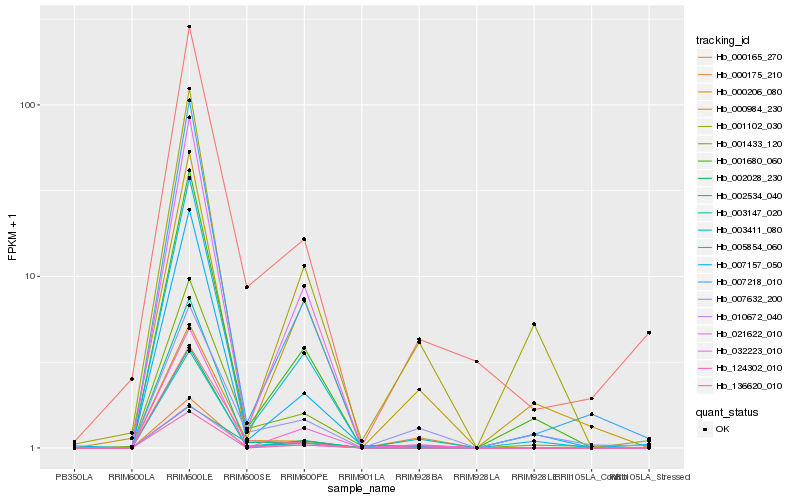
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010672_040 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_124302_010 |
0.1112097801 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 3 |
Hb_003147_020 |
0.1280335657 |
- |
- |
hypothetical protein POPTR_0019s07690g [Populus trichocarpa] |
| 4 |
Hb_002534_040 |
0.1328754862 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 5 |
Hb_032223_010 |
0.144813212 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_000165_270 |
0.1462896073 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 7 |
Hb_001433_120 |
0.1470321245 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 8 |
Hb_007632_200 |
0.147952157 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 9 |
Hb_000206_080 |
0.1564461362 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 10 |
Hb_136620_010 |
0.1569001305 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 11 |
Hb_003411_080 |
0.1646427111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005854_060 |
0.1646866262 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_002028_230 |
0.1694444618 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 14 |
Hb_001102_030 |
0.171094993 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_000984_230 |
0.1725250154 |
- |
- |
- |
| 16 |
Hb_000175_210 |
0.1732922782 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 17 |
Hb_007157_050 |
0.1739353442 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 18 |
Hb_007218_010 |
0.173999667 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 19 |
Hb_001680_060 |
0.1757313718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_021622_010 |
0.1770606722 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |