| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
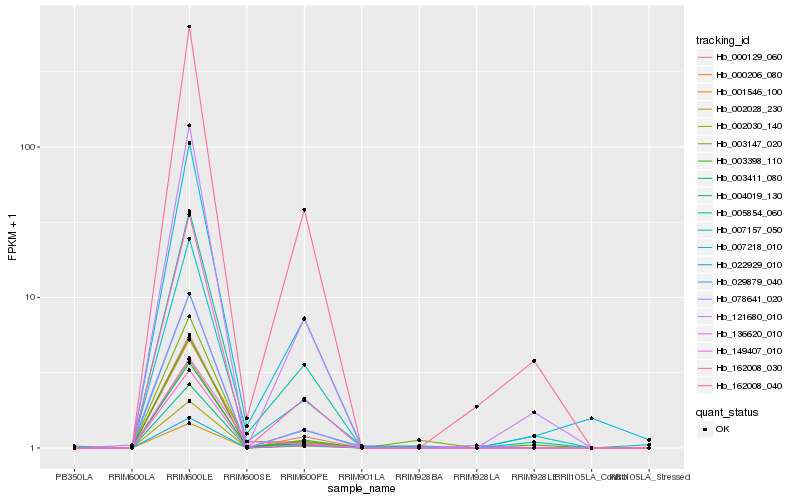
Hb_003411_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002028_230 |
0.0397865046 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 3 |
Hb_000206_080 |
0.0533837999 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 4 |
Hb_149407_010 |
0.0636017777 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 5 |
Hb_162008_030 |
0.0666595912 |
- |
- |
cytochrome p450 [Ageratina adenophora] |
| 6 |
Hb_000129_060 |
0.0735867071 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 7 |
Hb_029879_040 |
0.0744636493 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 8 |
Hb_004019_130 |
0.0766193487 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 9 |
Hb_136620_010 |
0.0768357444 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 10 |
Hb_121680_010 |
0.0915185278 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 11 |
Hb_078641_020 |
0.0937762664 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 12 |
Hb_005854_060 |
0.0944439586 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_007157_050 |
0.0968535564 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 14 |
Hb_007218_010 |
0.0990550617 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 15 |
Hb_001546_100 |
0.0992565382 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 16 |
Hb_162008_040 |
0.1004212314 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 17 |
Hb_003398_110 |
0.1025220767 |
- |
- |
PREDICTED: uncharacterized protein LOC105647583 [Jatropha curcas] |
| 18 |
Hb_002030_140 |
0.1038781989 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 19 |
Hb_003147_020 |
0.104223378 |
- |
- |
hypothetical protein POPTR_0019s07690g [Populus trichocarpa] |
| 20 |
Hb_022929_010 |
0.1049989323 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |