| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
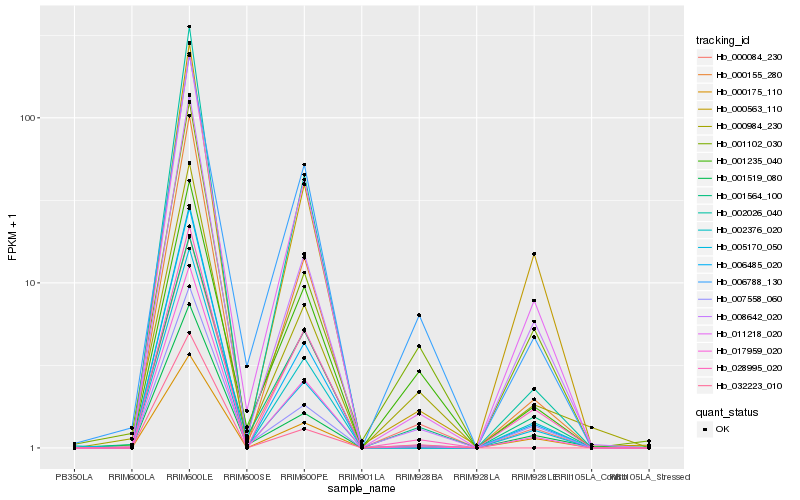
Hb_000984_230 |
0.0 |
- |
- |
- |
| 2 |
Hb_001102_030 |
0.0888792956 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_006788_130 |
0.1032075971 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 4 |
Hb_000084_230 |
0.1080115245 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 5 |
Hb_005170_050 |
0.1083729539 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 6 |
Hb_001235_040 |
0.1084735174 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 7 |
Hb_008642_020 |
0.1098845936 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 8 |
Hb_017959_020 |
0.111015038 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 9 |
Hb_011218_020 |
0.112466305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007558_060 |
0.1140750202 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 11 |
Hb_001519_080 |
0.1145820698 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_001564_100 |
0.1150681292 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 13 |
Hb_000175_110 |
0.1155867658 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 14 |
Hb_006485_020 |
0.1214994487 |
- |
- |
hypothetical protein JCGZ_15092 [Jatropha curcas] |
| 15 |
Hb_000155_280 |
0.1224284281 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45950 [Jatropha curcas] |
| 16 |
Hb_028995_020 |
0.1233519552 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 17 |
Hb_000563_110 |
0.1255236514 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 18 |
Hb_032223_010 |
0.1258440263 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 19 |
Hb_002376_020 |
0.1275353925 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 20 |
Hb_002026_040 |
0.1300076935 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |