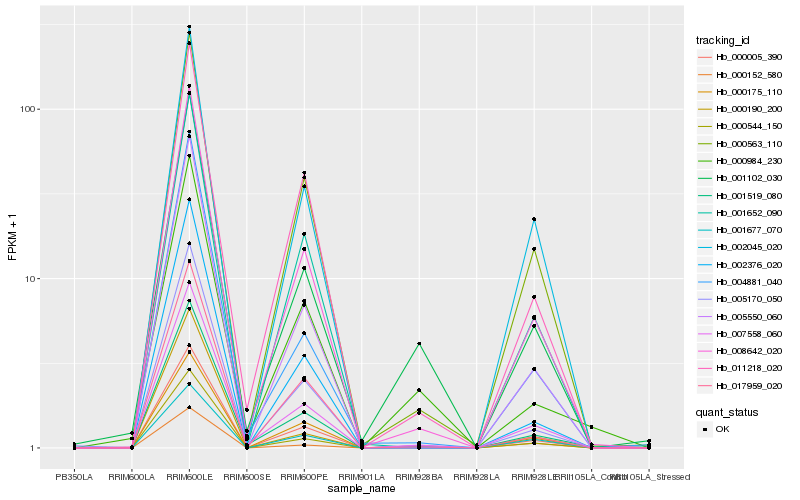
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_030 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_007558_060 |
0.0868850603 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 3 |
Hb_000984_230 |
0.0888792956 |
- |
- |
- |
| 4 |
Hb_008642_020 |
0.0961173395 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 5 |
Hb_005170_050 |
0.1058220925 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Populus euphratica] |
| 6 |
Hb_000175_110 |
0.1067691211 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 7 |
Hb_001519_080 |
0.11009714 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_000152_580 |
0.1115416153 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 9 |
Hb_004881_040 |
0.1121194375 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 10 |
Hb_000563_110 |
0.113232744 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 11 |
Hb_001677_070 |
0.1137470182 |
- |
- |
PREDICTED: putative caffeoyl-CoA O-methyltransferase At1g67980 [Jatropha curcas] |
| 12 |
Hb_005550_060 |
0.1181228971 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000544_150 |
0.1189630349 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 14 |
Hb_000005_390 |
0.1210019079 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 15 |
Hb_000190_200 |
0.1253350016 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 16 |
Hb_002376_020 |
0.1275620759 |
- |
- |
lectin, partial [Lathyrus palustris] |
| 17 |
Hb_011218_020 |
0.1294804261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_017959_020 |
0.1319786652 |
- |
- |
hypothetical protein POPTR_0011s03440g [Populus trichocarpa] |
| 19 |
Hb_001652_090 |
0.1320010296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002045_020 |
0.1328868363 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |