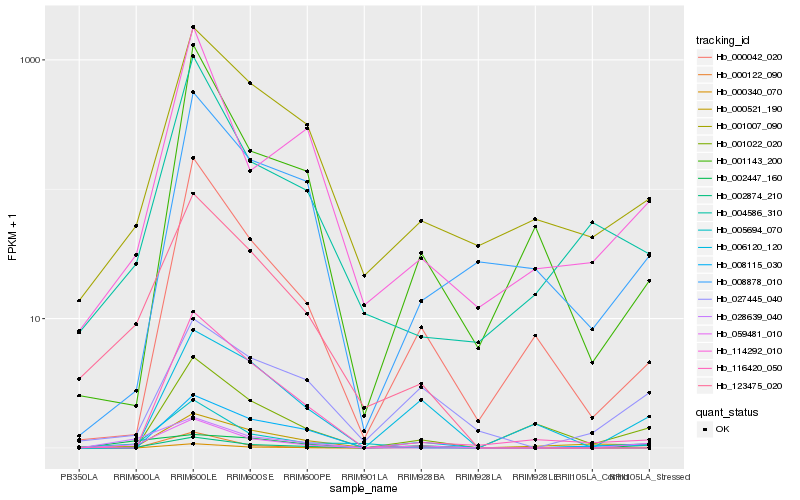
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_059481_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 2 |
Hb_002874_210 |
0.2151320126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001007_090 |
0.2358668204 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 4 |
Hb_005694_070 |
0.2370263884 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_123475_020 |
0.2405360555 |
- |
- |
- |
| 6 |
Hb_000042_020 |
0.2452496323 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002447_160 |
0.2562372402 |
- |
- |
- |
| 8 |
Hb_000122_090 |
0.257019256 |
- |
- |
hypothetical protein RCOM_0688100 [Ricinus communis] |
| 9 |
Hb_027445_040 |
0.2583554337 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 10 |
Hb_000521_190 |
0.2591048155 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 11 |
Hb_004586_310 |
0.2596431291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008115_030 |
0.2636235872 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 13 |
Hb_114292_010 |
0.2672185349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_116420_050 |
0.2698864346 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 15 |
Hb_006120_120 |
0.2699378705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000340_070 |
0.2712678353 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 17 |
Hb_028639_040 |
0.2717483794 |
- |
- |
PREDICTED: uncharacterized protein LOC105761689 [Gossypium raimondii] |
| 18 |
Hb_001022_020 |
0.2723225491 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 19 |
Hb_008878_010 |
0.2738328542 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 20 |
Hb_001143_200 |
0.2744261298 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |