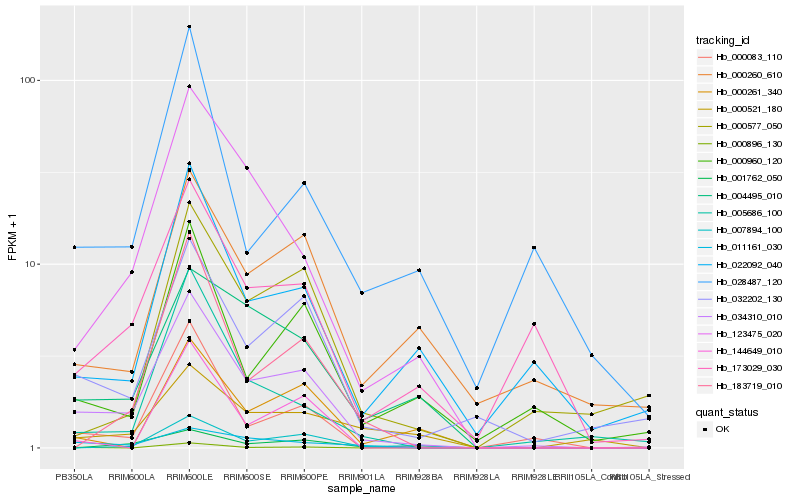
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034310_010 |
0.0 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_032202_130 |
0.1740588976 |
- |
- |
PREDICTED: uncharacterized protein LOC105643062 [Jatropha curcas] |
| 3 |
Hb_000083_110 |
0.1818072685 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 4 |
Hb_123475_020 |
0.1893542036 |
- |
- |
- |
| 5 |
Hb_011161_030 |
0.2005013318 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 6 |
Hb_173029_030 |
0.2047765013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004495_010 |
0.20951664 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 8 |
Hb_000960_120 |
0.2114773793 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 9 |
Hb_022092_040 |
0.2145591732 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 10 |
Hb_183719_010 |
0.21508693 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 11 |
Hb_001762_050 |
0.2175405418 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 12 |
Hb_005686_100 |
0.2190570273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000896_130 |
0.2209541681 |
transcription factor |
TF Family: CPP |
cysteine-rich polycomb-like family protein [Populus trichocarpa] |
| 14 |
Hb_007894_100 |
0.2216662513 |
- |
- |
PREDICTED: uncharacterized protein LOC105632952 [Jatropha curcas] |
| 15 |
Hb_144649_010 |
0.223761113 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 16 |
Hb_000260_610 |
0.2283528942 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 17 |
Hb_000577_050 |
0.2285624883 |
- |
- |
- |
| 18 |
Hb_000261_340 |
0.2347938405 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_000521_180 |
0.2386336183 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 20 |
Hb_028487_120 |
0.2415988007 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |