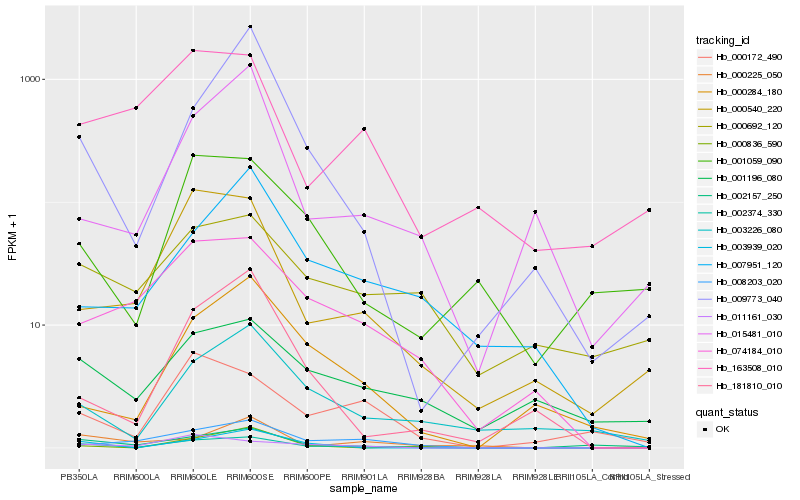
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_330 |
0.0 |
- |
- |
- |
| 2 |
Hb_002157_250 |
0.2161013467 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 3 |
Hb_074184_010 |
0.2259859695 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 4 |
Hb_008203_020 |
0.2355733972 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000540_220 |
0.2355920139 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 6 |
Hb_003939_020 |
0.2404513421 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 7 |
Hb_011161_030 |
0.2408019804 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 8 |
Hb_003226_080 |
0.2504613773 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 9 |
Hb_000172_490 |
0.2562899706 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 10 |
Hb_009773_040 |
0.2601959304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001196_080 |
0.2624493345 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 12 |
Hb_181810_010 |
0.2641820002 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 13 |
Hb_000836_590 |
0.2675096057 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 14 |
Hb_000284_180 |
0.269684902 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 15 |
Hb_163508_010 |
0.269950836 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 16 |
Hb_015481_010 |
0.2704080083 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 17 |
Hb_000225_050 |
0.2709584979 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_007951_120 |
0.2714221612 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 19 |
Hb_000692_120 |
0.2714763196 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 20 |
Hb_001059_090 |
0.2725581444 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |