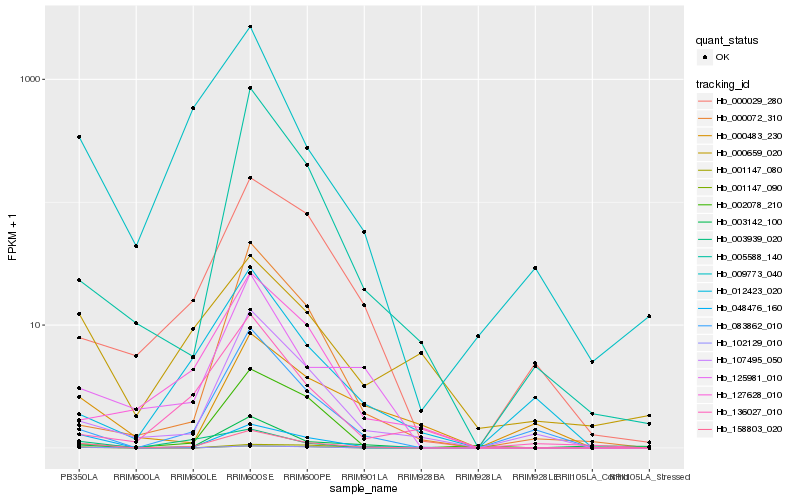
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158803_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_12890 [Jatropha curcas] |
| 2 |
Hb_048476_160 |
0.1761482843 |
- |
- |
hypothetical protein JCGZ_19593 [Jatropha curcas] |
| 3 |
Hb_083862_010 |
0.2133954102 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 4 |
Hb_107495_050 |
0.2268805294 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_125981_010 |
0.2271865886 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 6 |
Hb_102129_010 |
0.2296717836 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 7 |
Hb_009773_040 |
0.2307024246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001147_090 |
0.242890441 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 9 |
Hb_003142_100 |
0.2440150461 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Eucalyptus grandis] |
| 10 |
Hb_003939_020 |
0.2481182861 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 11 |
Hb_000029_280 |
0.2542700904 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 12 |
Hb_001147_080 |
0.2574946353 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_127628_010 |
0.2582909215 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_005588_140 |
0.2585695574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012423_020 |
0.2665708594 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 16 |
Hb_000659_020 |
0.2672323276 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 17 |
Hb_000072_310 |
0.2698442973 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 18 |
Hb_002078_210 |
0.2712348559 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 19 |
Hb_136027_010 |
0.2715262712 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_000483_230 |
0.2722739836 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |