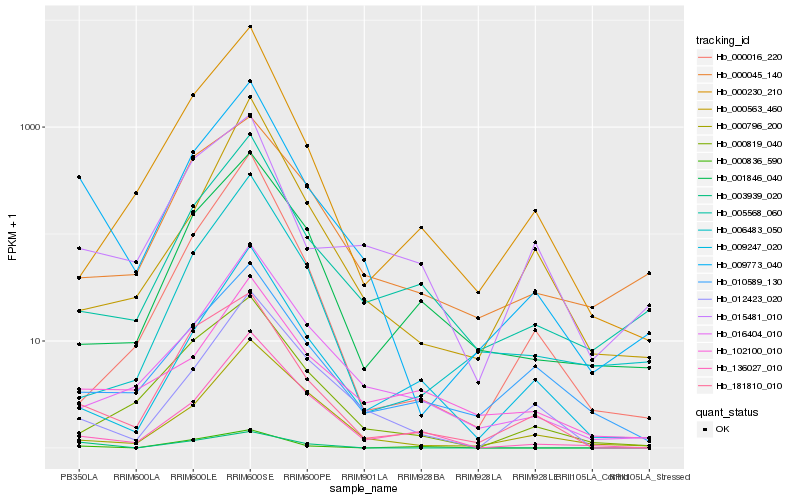
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009773_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_181810_010 |
0.1807442613 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 3 |
Hb_005568_060 |
0.1943572269 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 4 |
Hb_016404_010 |
0.1956707494 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_003939_020 |
0.2059716573 |
- |
- |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 6 |
Hb_012423_020 |
0.2062200101 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 7 |
Hb_136027_010 |
0.2067778349 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_000796_200 |
0.2104181921 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 9 |
Hb_000230_210 |
0.2121067051 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 10 |
Hb_000016_220 |
0.2122322915 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 11 |
Hb_000819_040 |
0.2141068948 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_102100_010 |
0.2144371662 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_009247_020 |
0.2164917534 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 14 |
Hb_015481_010 |
0.2165359165 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 15 |
Hb_001846_040 |
0.2174029642 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_010589_130 |
0.2189299719 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_000563_460 |
0.2212415253 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 18 |
Hb_000045_140 |
0.2217406157 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 19 |
Hb_006483_050 |
0.2221334255 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 20 |
Hb_000836_590 |
0.2263885512 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |