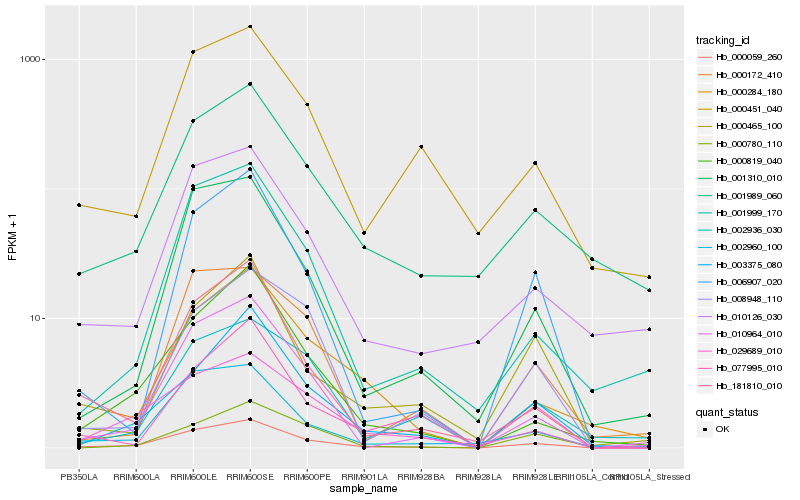
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_260 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 2 |
Hb_029689_010 |
0.132679465 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_010964_010 |
0.1363087072 |
- |
- |
- |
| 4 |
Hb_002960_100 |
0.139719304 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 5 |
Hb_000819_040 |
0.1484470252 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001310_010 |
0.1493364512 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 7 |
Hb_181810_010 |
0.150252715 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 8 |
Hb_003375_080 |
0.152770513 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006907_020 |
0.1581402573 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 10 |
Hb_000451_040 |
0.1601829536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001989_060 |
0.1610449941 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000284_180 |
0.1618134979 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 13 |
Hb_010126_030 |
0.1639473171 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001999_170 |
0.1642158449 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 15 |
Hb_008948_110 |
0.1686256318 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 16 |
Hb_002936_030 |
0.1714314409 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000780_110 |
0.1746304909 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 18 |
Hb_000172_410 |
0.1750614316 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_077995_010 |
0.1784465614 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_000465_100 |
0.1792767779 |
- |
- |
catalytic, putative [Ricinus communis] |