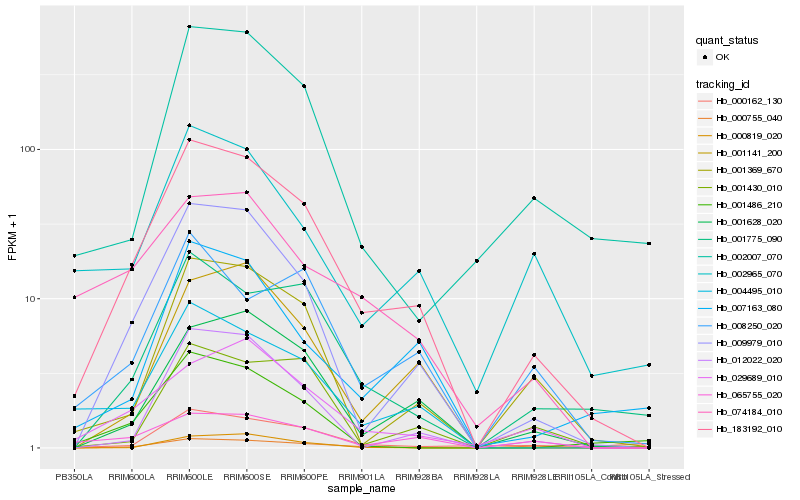
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183192_010 |
0.0 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 2 |
Hb_009979_010 |
0.1209805738 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_004495_010 |
0.1561737971 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 4 |
Hb_001628_020 |
0.1595385969 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_001369_670 |
0.170386556 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_029689_010 |
0.1732747908 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_012022_020 |
0.1831674905 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 8 |
Hb_001775_090 |
0.1848064686 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 9 |
Hb_001141_200 |
0.1889138283 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 10 |
Hb_007163_080 |
0.1926092263 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 11 |
Hb_065755_020 |
0.1928365316 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 12 |
Hb_001486_210 |
0.1933700414 |
- |
- |
- |
| 13 |
Hb_002007_070 |
0.1977799995 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 14 |
Hb_000755_040 |
0.1988157293 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 15 |
Hb_002965_070 |
0.2008084972 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 16 |
Hb_000162_130 |
0.2021954987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_074184_010 |
0.2023146552 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 18 |
Hb_000819_020 |
0.2034802868 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 19 |
Hb_001430_010 |
0.2061165672 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_008250_020 |
0.2083905848 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |