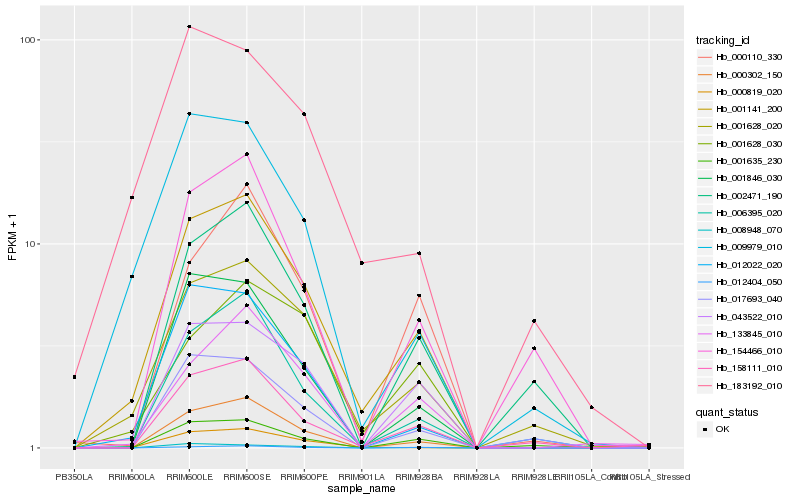
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_200 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 2 |
Hb_001628_020 |
0.1141697056 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 3 |
Hb_006395_020 |
0.1528669273 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 4 |
Hb_012404_050 |
0.1551818708 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 5 |
Hb_043522_010 |
0.1607244251 |
- |
- |
- |
| 6 |
Hb_158111_010 |
0.1644112161 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 7 |
Hb_000819_020 |
0.1676636149 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 8 |
Hb_009979_010 |
0.1680969946 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_001846_030 |
0.168740692 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 10 |
Hb_000302_150 |
0.1690542316 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 11 |
Hb_133845_010 |
0.1728443221 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_012022_020 |
0.1731804592 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 13 |
Hb_000110_330 |
0.1798114965 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 14 |
Hb_001628_030 |
0.1815185555 |
- |
- |
PREDICTED: uncharacterized protein LOC105629266 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002471_190 |
0.1842903231 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 16 |
Hb_154466_010 |
0.1873401334 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_017693_040 |
0.1875323129 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 18 |
Hb_008948_070 |
0.1879975179 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001635_230 |
0.1885870375 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_183192_010 |
0.1889138283 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |