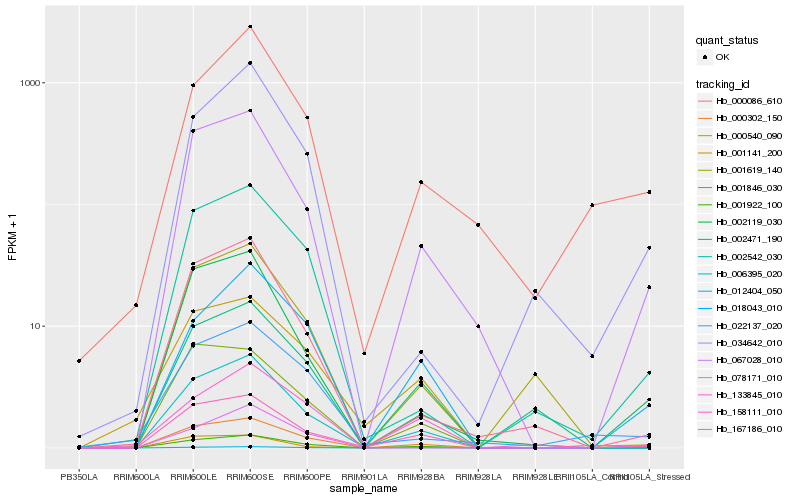
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001922_100 |
0.0 |
- |
- |
S-type anion channel SLAH2 [Triticum urartu] |
| 2 |
Hb_000302_150 |
0.048210123 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 3 |
Hb_067028_010 |
0.1076986973 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 4 |
Hb_018043_010 |
0.1281568753 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 5 |
Hb_006395_020 |
0.1381922062 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 6 |
Hb_158111_010 |
0.1445961044 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 7 |
Hb_002119_030 |
0.1540755042 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 8 |
Hb_002542_030 |
0.1617518378 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 9 |
Hb_012404_050 |
0.1634453055 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 10 |
Hb_133845_010 |
0.1685177297 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_167186_010 |
0.1798066903 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 12 |
Hb_001846_030 |
0.1804906692 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 13 |
Hb_000540_090 |
0.1821959619 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 14 |
Hb_000086_610 |
0.18553158 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 15 |
Hb_022137_020 |
0.1858957099 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 16 |
Hb_001619_140 |
0.1890962525 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_001141_200 |
0.1894336515 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 18 |
Hb_078171_010 |
0.1906772369 |
- |
- |
PREDICTED: uncharacterized RING finger protein P8B7.15c-like [Jatropha curcas] |
| 19 |
Hb_002471_190 |
0.1910658054 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 20 |
Hb_034642_010 |
0.1917891699 |
- |
- |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |