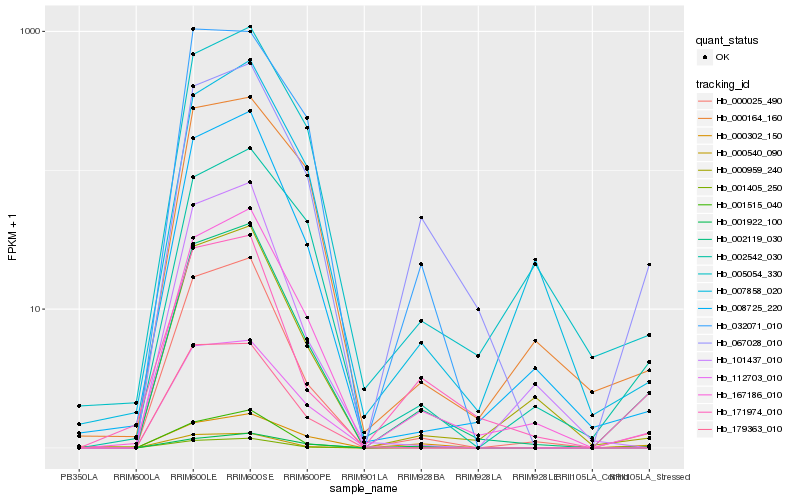
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002119_030 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 2 |
Hb_000025_490 |
0.0807833561 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 3 |
Hb_067028_010 |
0.0911980169 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 4 |
Hb_167186_010 |
0.0933447373 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 5 |
Hb_008725_220 |
0.118871475 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 6 |
Hb_000959_240 |
0.1229082318 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_005054_330 |
0.1257712426 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 8 |
Hb_171974_010 |
0.1306325186 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 9 |
Hb_001405_250 |
0.1336001238 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 10 |
Hb_001515_040 |
0.1400203094 |
- |
- |
- |
| 11 |
Hb_002542_030 |
0.1424432484 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 12 |
Hb_000302_150 |
0.1429700722 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 13 |
Hb_007858_020 |
0.1464466066 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 14 |
Hb_032071_010 |
0.1464673392 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 15 |
Hb_179363_010 |
0.146902558 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 16 |
Hb_000540_090 |
0.1475352122 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 17 |
Hb_000164_160 |
0.1515583718 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_112703_010 |
0.1525688237 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 19 |
Hb_001922_100 |
0.1540755042 |
- |
- |
S-type anion channel SLAH2 [Triticum urartu] |
| 20 |
Hb_101437_010 |
0.1566037375 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |