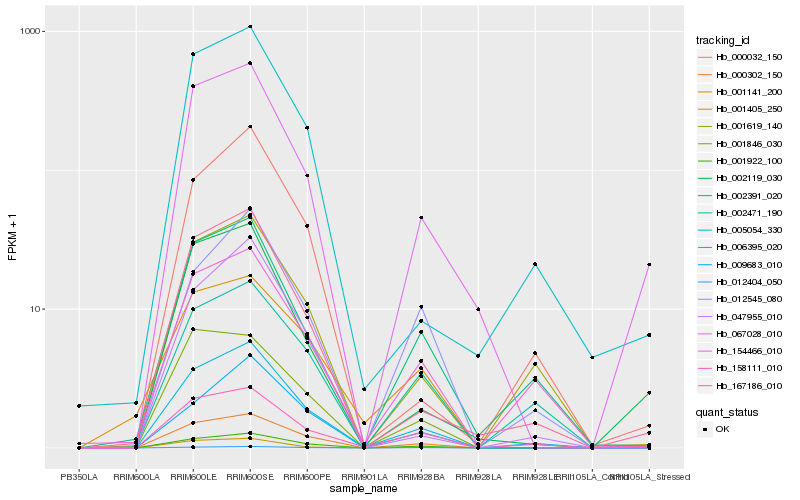
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006395_020 |
0.0 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 2 |
Hb_000302_150 |
0.1150438759 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 3 |
Hb_001405_250 |
0.1203496606 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 4 |
Hb_067028_010 |
0.1261756833 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 5 |
Hb_012404_050 |
0.1290800167 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 6 |
Hb_167186_010 |
0.1300981951 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 7 |
Hb_001922_100 |
0.1381922062 |
- |
- |
S-type anion channel SLAH2 [Triticum urartu] |
| 8 |
Hb_001846_030 |
0.1397377484 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 9 |
Hb_158111_010 |
0.1398388649 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 10 |
Hb_047955_010 |
0.1417601752 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 11 |
Hb_001619_140 |
0.142870698 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_002391_020 |
0.1462700421 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 13 |
Hb_009683_010 |
0.147474842 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_001141_200 |
0.1528669273 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 15 |
Hb_154466_010 |
0.1530962165 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_012545_080 |
0.1540706852 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 17 |
Hb_005054_330 |
0.1548296855 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 18 |
Hb_000032_150 |
0.1574777665 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 19 |
Hb_002119_030 |
0.1586004788 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 20 |
Hb_002471_190 |
0.1591694737 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |