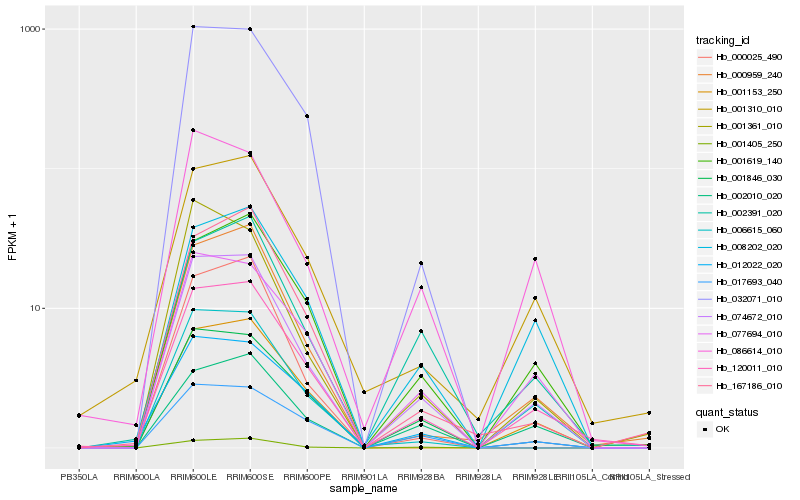
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074672_010 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_120011_010 |
0.0852620389 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 3 |
Hb_077694_010 |
0.1052239535 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 4 |
Hb_006615_060 |
0.1088443846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000959_240 |
0.1188297336 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_017693_040 |
0.1209252897 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 7 |
Hb_001619_140 |
0.1218920267 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_001405_250 |
0.1235667859 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 9 |
Hb_086614_010 |
0.127192076 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002391_020 |
0.1296931503 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 11 |
Hb_001361_010 |
0.1309785717 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 12 |
Hb_008202_020 |
0.1318733839 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_012022_020 |
0.1328266178 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 14 |
Hb_002010_020 |
0.1341400792 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 15 |
Hb_032071_010 |
0.1359671854 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 16 |
Hb_001153_250 |
0.1369265132 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 17 |
Hb_001846_030 |
0.1375810651 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 18 |
Hb_001310_010 |
0.1399354816 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 19 |
Hb_167186_010 |
0.1409235266 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 20 |
Hb_000025_490 |
0.1430715965 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |