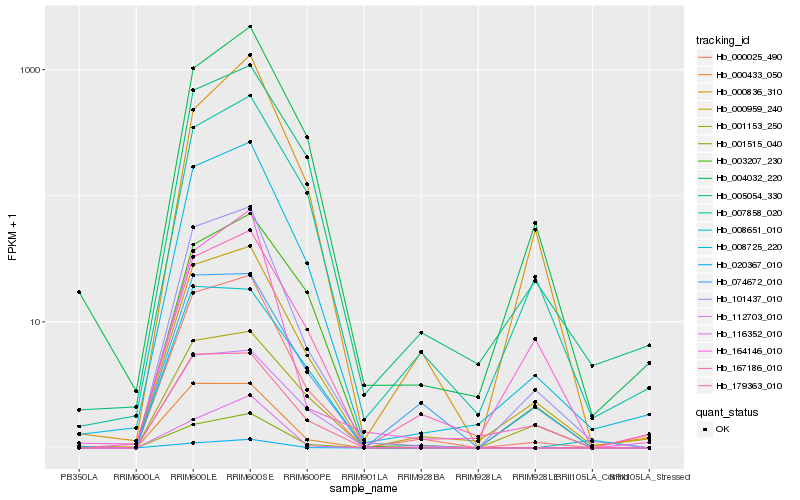
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101437_010 |
0.0 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 2 |
Hb_008725_220 |
0.0776814694 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 3 |
Hb_000959_240 |
0.0824817672 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001515_040 |
0.0919476624 |
- |
- |
- |
| 5 |
Hb_000025_490 |
0.0965466394 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 6 |
Hb_020367_010 |
0.1032652795 |
- |
- |
hypothetical protein PRUPE_ppa004271mg [Prunus persica] |
| 7 |
Hb_000433_050 |
0.1171348094 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 8 |
Hb_179363_010 |
0.1271922169 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_004032_220 |
0.1280752115 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 10 |
Hb_007858_020 |
0.1287639185 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 11 |
Hb_167186_010 |
0.1300139598 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 12 |
Hb_005054_330 |
0.1310068584 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 13 |
Hb_001153_250 |
0.1368113125 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 14 |
Hb_000836_310 |
0.1430766118 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 15 |
Hb_074672_010 |
0.1443146323 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 16 |
Hb_112703_010 |
0.1481312971 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 17 |
Hb_116352_010 |
0.149490779 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_003207_230 |
0.1519193088 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 19 |
Hb_008651_010 |
0.1550781998 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_164146_010 |
0.1564401774 |
- |
- |
- |