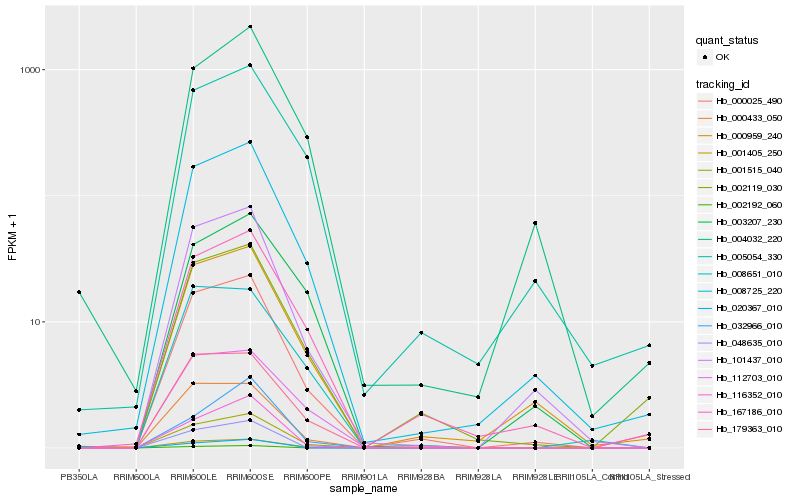
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001515_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_020367_010 |
0.0570137141 |
- |
- |
hypothetical protein PRUPE_ppa004271mg [Prunus persica] |
| 3 |
Hb_008725_220 |
0.0846241997 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 4 |
Hb_000025_490 |
0.091407209 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 5 |
Hb_101437_010 |
0.0919476624 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 6 |
Hb_179363_010 |
0.1054755234 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_000433_050 |
0.1057343059 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 8 |
Hb_116352_010 |
0.1163895803 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_167186_010 |
0.1237394528 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 10 |
Hb_112703_010 |
0.1241309488 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 11 |
Hb_000959_240 |
0.124891426 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004032_220 |
0.1396675897 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 13 |
Hb_032966_010 |
0.139737005 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_002119_030 |
0.1400203094 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 15 |
Hb_005054_330 |
0.1421680337 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 16 |
Hb_008651_010 |
0.1432056848 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001405_250 |
0.1444791015 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 18 |
Hb_003207_230 |
0.1457393499 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 19 |
Hb_002192_060 |
0.1495607901 |
- |
- |
- |
| 20 |
Hb_048635_010 |
0.1496022618 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |